 IVD Raw Materials
IVD Raw Materials

Home » Oligo Synthesis Applications » Small Nucleic Acid Drug Synthesis
With over 22 years of expertise, GenScript offers comprehensive synthesis services for small nucleic acid drug, including custom siRNA, ASO, miRNA, aptamer, tRNA, and saRNA(small activating RNA). These services span from R&D to cGMP production, supporting small RNA drug development.
Superior Quality, Ready in 4 Days
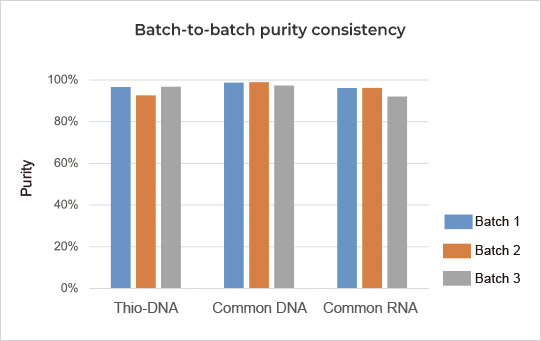
siRNA/ASO products with ≥90% purity and excellent consistency
AI-Enhanced Precision Design
AI combined with experimental validation optimizes design logic
Proprietary Modifications
Boost drug development with enhanced stability and reduced off-target effects
| Service Type | siRNA | ASO |
|---|---|---|
| Length | 20-30nt | 15-30nt |
| Quantity | 2 nmol to Kg level | 5 nmol to Kg level |
| Type |
|
|
| Purification | RNase free HPLC, desalt | |
| Modification | Locked Nucleic Acid (LNA), thioate, methylation, 2-Ome etc. (~200 types) | |
| Conjugation | Antibody, peptide, GalNAc, cholesterol etc. | |
Genscript also offers custom synthesis services for miRNA, aptamer, tRNA, and saRNA. Contact us for more information or to request a quote
| QC Item | Test Method | Specification | Standard QC | Customized QC |
|---|---|---|---|---|
| Appearance | Visual Inspect | White to Pale Yellow Powder | ||
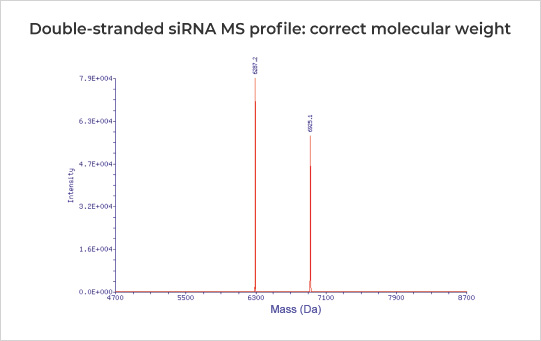
| Identity by MW | Mass Spectrometry | Theoretical Average MW ± 0.1% | ||
| LC-MS | Liquid Chromatography–mass Spectrometry | Report Result | ||
| LC MS/MS Sequencing | High-resolution Mass Spectrometry Sequencing | 100% Sequence Coverage | ||
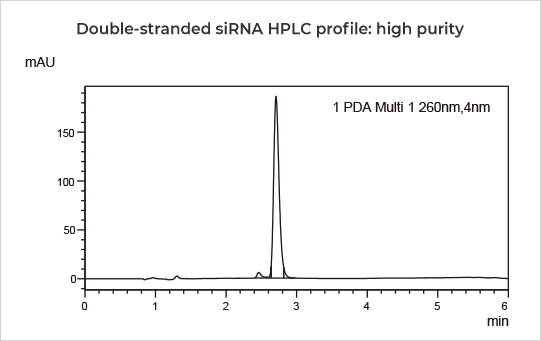
| Purity by HPLC | High Performance Liquid Chromatography | Double-strand siRNA ≥ 90% | ||
| Purity by CGE | Capillary Gel Electrophoresis | Report Result | ||
| Purity by PAGE | Polyacrylamide Gel Electrophoresis | Report Result | ||
| pH Measurement | Micro pH Electrode Method | Report Result | ||
| TEA Salt Residue Detection | Gas Chromatography-mass Spectrometry Measurement | Report Result | ||
| Solvent Residue | Gas Chromatography-mass Spectrometry Measurement | Report Result | ||
| Endotoxin | Chromogenic Method | Report Result | ||
| Bacterial Testing | Direct Inoculation / Membrane Filter Method | Report Result |
Please find more customized QC information in Oligo Quality Control Service>>.
siRNA Guarantee Package: AI-Enhanced Precision Design, ensuring knockdown efficiency ≥80%
GenScript has developed seven modification combinations (GS1-GS7) through bioinformatics screening and experimental selection. These strategies include specific modifications in type, number, and position to improve siRNA efficacy. AI-assisted predictions further accelerate selection, saving time and cost.
Preparation (1 Day)
Provide candidate siRNA sequences.
Design (2-3 Days)
Bioinformatic prediction of knockdown efficiency.
Synthesis (4+ Days)
Select and synthesize modified siRNA.
Validation (Optional)
Test actual knockdown efficiency, off-target effects, and stability.
Note: The effectiveness of modification combinations for off-target reduction and stability varies by siRNA sequence. We recommend first identifying unmodified candidates with strong knockdown efficiency, then testing test multiple predicted high-efficiency modification combinations for knockdown efficiency, off-target effects, and stability.
* GenScript offers a free siRNA design tool utilizing classic design logic. If you prefer an AI-enhanced design platform, please choose the siRNA guarantee package or send a request to oligo@genscript.com.
Need help from an expert?




20 nt double-stranded siRNA with 29 2'-O-Methyl modifications, 3 phosphothioate groups, 13 2'-Fluoro modifications, and 1 GalNAc modification, exhibited purity up to 96.55%



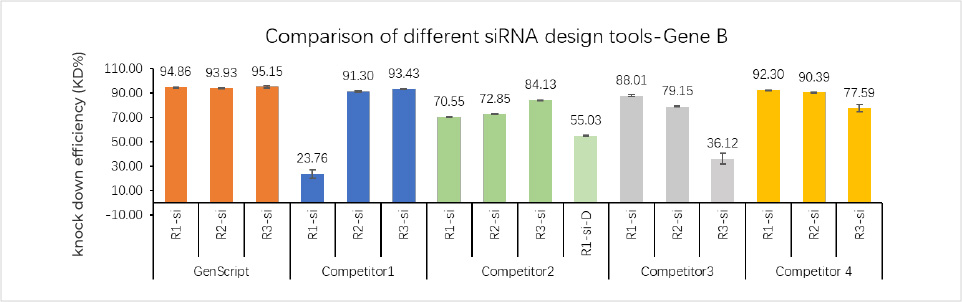
siRNA sequences designed for Gene B with GenScript's bioinformatic design platform achieved higher knockdown efficiency (KD%) than those created with other vendors' tools.
Case 1: Proprietary modifications significantly enhance siRNA stability.
Method: For different modification schemes, the metabolic rate and metabolites in a serum environment were analyzed to select the most stable modification schemes for subsequent optimization and development.
Results

Case 2: Proprietary modifications significantly reduce siRNA off-target effects.
Method
RNA-Seq was used to detect and enumerate DEGs(differential expression genes) following treatment with both modified and unmodified siRNA, assessing the extent of mRNA upregulation or downregulation in off-target genes.
Results
Table 1: Modified siRNA (siRNA93-GS) significantly reduced the number of DEGs compared with unmodified siRNA.
| Target Gene | Sample | RNA-Seq Detection | Type I: Homologous off-target genes prediction | Type II: miRNA-like similar off-target genes prediction | ||||
|---|---|---|---|---|---|---|---|---|
| DEGs | AS_blast | SS_blast | AS_mirna_off-target | SS_mirna_off-target | Total mirna_off-target | Down regulated off-target | ||
| HAO1 | siRNA93 | 58 | 0 | 0 | 26 | 6 | 28 | 23 |
| siRNA93-GS | 7 | 0 | 0 | 1 | 0 | 1 | 1 | |
Off-Target Effects of Unmodified siRNA - Gene Transcription Changes

Off-Target Effects of GenScript’s Proprietary Modified siRNA – Gene Transcription Changes

Case 3: siRNA modification combinations achieve a 27-fold IC50 reduction compared with commercial options
Method:
qPCR was used to measure IC50 of siRNAs with identical sequences but different modifications at the mRNA level.
Results:
Compared with the commercial siRNA modification scheme (Vutrisiran), GenScript's GS1-6 modifications significantly lowered IC50. GS5 reduced IC50 by up to 27-fold, enabling effective knockdown at much lower doses.
| Samples | IC50 (nM) | Knockdown(%)-10nM |
|---|---|---|
| TTR_siRNA_naked | 0.00257 | 97.93% |
| TTR_siRNA_GS5 | 0.00517 | 98.05% |
| TTR_siRNA_GS3 | 0.00704 | 97.35% |
| TTR_siRNA_GS6 | 0.0133 | 97.34% |
| TTR_siRNA_GS2 | 0.0149 | 97.77% |
| TTR_siRNA_GS4 | 0.0553 | 97.37% |
| TTR_siRNA_GS1 | 0.114 | 95.50% |
| TTR_siRNA_Vutrisiran | 0.142 | 94.95% |
| TTR_siRNA_GS7 | 0.225 | 91.46% |
IC50 Assessment Protocol: HepG2 cells were transfected with unmodified or modified siRNA targeting the TTR site using RNAiMAX across a 10-dose gradient (100 nM to 0.0000512 nM). RNA was extracted after 48 hours, reverse transcribed, and analyzed by qPCR to determine mRNA knockdown efficiency and IC50 values.

siRNA Lipofection Protocol
Free Download