Methods: HEK293T cells (0.2 million cells) were transfected
with 1 μg PE2/PE3
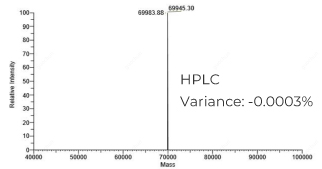
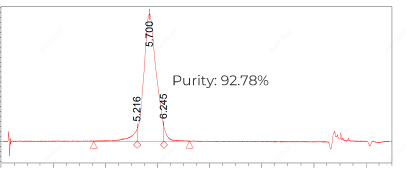
mRNA, 30pmol nicking sgRNA and 90pmol pegRNA of different lengths (HPLC grade) targeting the
HEK3 gene, via electroporation. Genome DNA was then extracted 3 days post-transfection for
editing efficiency analysis via Sanger sequencing.
Results: pegRNA with a tevopreQ1 motif achieved
editing efficiencies as
high as 43.4%, which was significantly higher than that of the pegRNA without
the tevopreQ1
motif.
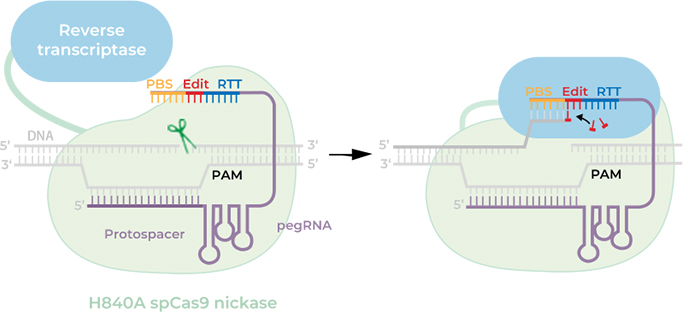
Note: the 218 nt pegRNA is an extended version of 181 nt with a
tevopreQ1 motif
(37 nt) at the 3' end. It has been reported that adding the RNA structural motifs evopreQ1
or
mpknot to the 3' end of a pegRNA can protect the 3' end from exonuclease digestion (Nelson,
J. W. et al., 2022).