-
REAGENT SERVICES
Hot!
-
Most Popular Services
-
Molecular Biology
-
Recombinant Antibody/Protein
-
Reagent Antibody
-
CRISPR Gene Editing
-
DNA Mutant Library
-
IVT RNA and LNP Formulations
-
Oligo Synthesis
-
Peptides
-
Cell Engineering
-
- Gene Synthesis FLASH Gene
- GenBrick™ Up to 200kb
- Gene Fragments Up to 3kb now
- Plasmid DNA Preparation Upgraded
- Cloning and Subcloning
- ORF cDNA Clones
- mRNA Plasmid Solutions New!
- Cell free mRNA Template New!
- AAV Plasmid Solutions New!
- Mutagenesis
- GenCircle™ Double-Stranded DNA New!
- GenSmart™ Online Tools
-
-
PRODUCTS
-
Most Popular Reagents
-
 Instruments
Instruments
-
Antibodies
-
ELISA Kits
-
Protein Electrophoresis and Blotting
-
Protein and Antibody Purification
-
Recombinant Proteins
-
Molecular Biology
-
Stable Cell Lines
-
Cell Isolation and Activation
-
 IVD Raw Materials
IVD Raw Materials
-
 Therapy Applications
Therapy Applications
-
Resources
-
- All Instruments
- Automated Protein and Antibody Purification SystemNew!
- Automated Plasmid MaxiprepHot!
- Automated Plasmid/Protein/Antibody Mini-scale Purification
- eBlot™ Protein Transfer System
- eStain™ Protein Staining System
- eZwest™ Lite Automated Western Blotting Device
- CytoSinct™ 1000 Cell Isolation Instrument
-
- Pharmacokinetics and Immunogenicity ELISA Kits
- Viral Titration QC ELISA Kits
- -- Lentivirus Titer p24 ELISA KitHot!
- -- MuLV Titer p30 ELISA KitNew!
- -- AAV2 and AAVX Titer Capsid ELISA Kits
- Residual Detection ELISA Kits
- -- T7 RNA Polymerase ELISA KitNew!
- -- BSA ELISA Kit, 2G
- -- Cas9 ELISA KitHot!
- -- Protein A ELISA KitHot!
- -- His tagged protein detection & purification
- dsRNA ELISA Kit
- Endonuclease ELISA Kit
- COVID-19 Detection cPass™ Technology Kits
-
- Automated Maxi-Plasmid PurificationHot!
- Automated Mini-Plasmid PurificationNew!
- PCR Reagents
- S.marcescens Nuclease Benz-Neburase™
- DNA Assembly GenBuilder™
- Cas9 / Cas12a / Cas13a Nucleases
- Base and Prime Editing Nucleases
- GMP Cas9 Nucleases
- CRISPR sgRNA Synthesis
- HDR Knock-in Template
- CRISPR Gene Editing Kits and Antibodies
-
![AmMag™ Quatro Automated Plasmid Purification]() AmMag™ Quatro automated plasmid purification
AmMag™ Quatro automated plasmid purification
-
![Anti-Camelid VHH]() MonoRab™ Anti-VHH Antibodies
MonoRab™ Anti-VHH Antibodies
-
![ELISA Kits]() ELISA Kits
ELISA Kits
-
![Precast Gels]() SurePAGE™ Precast Gels
SurePAGE™ Precast Gels
-
![Quatro ProAb Automated Protein and Antibody Purification System]() AmMag™ Quatro ProAb Automated Protein and Antibody Purification System
AmMag™ Quatro ProAb Automated Protein and Antibody Purification System
-
![Target Proteins]() Target Proteins
Target Proteins
-
![AmMag™ Quatro Automated Plasmid Purification]() AmMag™ Quatro automated plasmid purification
AmMag™ Quatro automated plasmid purification
-
![Stable Cell Lines]() Stable Cell Lines
Stable Cell Lines
-
![Cell Isolation and Activation]() Cell Isolation and Activation
Cell Isolation and Activation
-
 IVD Raw Materials
IVD Raw Materials
-
![Quick
Order]() Quick Order
Quick Order
-
![Quick
Order]() Quick Order
Quick Order
- APPLICATIONS
- RESOURCES
- ABOUT US
- SIGN IN My Account SIGN OUT
- REGISTER
Resources » Weekly Scientific Insight » A Guide to Efficient CRISPR gRNA Design: Principles and Design Tools
 doreen.ding
doreen.dingA Guide to Efficient CRISPR gRNA Design: Principles and Design Tools
Editor: Doreen Ding, Dr. Xia Sheng
CRISPR has rapidly become the most popular gene editing tool among academic researchers and industry alike due to its ease of design, usage and cost-effectiveness, relative to previous gene editing systems like TALENs or ZFNs. CRISPR-Cas systems can be easily engineered to target specific genes and produce edits by designing different guide RNA (gRNA).
To help those interested in designing their own CRISPR experiments, we’ve asked our in-house bioinformatic experts to share their experience regarding how to design effective CRISPR gRNAs and explain the principles behind GenScript’s design algorithms.
Basic Rules
Take the most commonly used CRISPR-Cas9 system as an example, a CRISPR-Cas9 system is composed of a Cas9 nuclease and a gRNA:
- A Cas9 protein first recognizes a specific protospacer adjacent motif (PAM) sequence in target sequence, before it binds and cuts at that sequence. There are different PAM sequences for different kinds of Cas nuclease. For example, "NGG" is recognized by SpCas9, “NNG" is recognized by scCas9, and "TTT(A/C/G)" is recognized by Cas12a.
- gRNA consists of a structural element called a "scaffold” or tracrRNA, which binds to the Cas9 protein, and a 20nt guiding element called "spacer" or crRNA which recognizes the target sequence by standard Watson-Crick base pairing. When these two elements are fused together in a single molecule, which is the most commonly used format, the term sgRNA (or single guide RNA) is used.
The basic goal in sgRNA design is to select a 20nt target sequence immediately upstream of a PAM site. The complementary 20nt spacer RNA directs the Cas9 nuclease to the specific genomic location to be edited. The target sequence should be unique within the genome to avoid off-target effects.
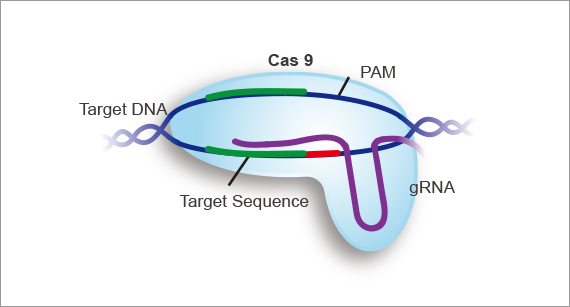
The design principles of sgRNA
Key Parameters
Here are some key parameters and evaluation logic to consider when designing your gRNA:
1. On-Target Efficiency: High on-target efficiency means the guide is predicted to have high editing efficiency of target site. Various algorithms have been developed to predict gRNA on-target efficiency, based on datasets from studies on the activities of thousands of gRNAs. Here we will introduce five commonly-used scoring methods:
① Rule Set:
- Reference: Developed by Doench et. al. [1] in 2014.
- Basis: This scoring algorithm is based on the knock-out efficiency data of 1,841 sgRNAs in actual experiments to classify which kinds of sgRNA works better. 30nt sequence including 20nt sgRNA bind area, 3nt PAM sequence and nearby sequences in the target sequence will all be taken into consideration.
- Scoring mode: A scoring matrix was then used to assign a score to each sgRNA. 80% of sgRNA with highest scores achieved high editing efficiency in actual experiments according to the author.
- Application: CHOPCHOP
② Rule Set 2:
- Reference: Updated by Doench's team in 2016 [2]
- Basis: The scoring algorithm is based on the knock-out efficiency data of 43,90 sgRNAs in actual experiments including 2,549 new gRNAs and previous 1,841 sgRNAs' dataset. The relationship between 30nt target sequence and editing efficiency is considered as well as Rule Set.
- Scoring mode: Gradient-boosted regression trees was used to assign a score to each sgRNA.
- Application: CHOPCHOP. CRISPOR
③ Rule set 3:
- Reference: Updated by Doench's team in 2022 [3].
- Basis: Trained on 7 existing gRNA efficiency dataset of 47k gRNAs in actual experiments. Rule set 3 takes the tracrRNA (scaffold for gRNA) into consideration. There are two types of logics as Hsu2013 and Chen2013 for different tracrRNA. Hsu2013 logics is recommended for any tracrRNA that have a T in the 5th position, such as tracrRNA sequence start with GTTTTAG.
- Scoring mode: The model was trained on Gradient Boosting framework rather than deep learning models, for faster training time.
- Application: GenScript, CRISpick.
④ CRISPRscan:
- Reference: Developed by Moreno-Mateos in 2015 [4].
- Basis: Predictive model based on the activity data of 1,280 gRNAs targeting 128 genes validated in vivo in zebra fish.
- Application: Chopchop, CRISPOR
⑤ Lindel:
- Reference: Developed by Chen in 2019[5].
- Basis: The author profiled ∼1.16 million mutation events resulting from Cas9-mediated cleavage and indel of 6872 synthetic target sequences in actual experiments.
- Score mode: A logistic regression model was developed to predict insertions and deletions that result from CRISPR/Cas9-mediated cleavage. Lindel uses a 60 bp sequence centered at the cleavage site as an input, and predicts a frameshift ratio. Compared with other indel predicting models, such as inDelphi and ForeCasT, Lindel is generally more accurate.
- Application: CRISPOR
2. Off-Target Risks: Specificity means avoiding off-target mutations, which could lead to unintended consequences. An sgRNA design should include a thorough genome-wide analysis of potential off-target sites that share significant homology with the target sequence. Here we will introduce 3 commonly-used scoring methods for off-target evaluation.
① Homology Analysis:
- Basis: Focuses on sequences similar to the given sgRNA in the genome. The more mismatches between designed sgRNA and off-target sequence there are, the lower off target effect there will be. Sequences that fit the PAM sequence (for example, "NGG") with fewer than three nucleotide mismatches are counted after a genome-wide search.
- Score mode: Sequences with only one mismatch imply high off-target potential, while sequences of two or three mismatches should be limited to as few as possible. Sequences with zero mismatches should be completely removed from consideration. The homology analysis can be improved by assigning different weights for mismatches at various positions. For example, mismatches closer to the PAM sequence can be assigned higher weights.
② MIT:
- Reference: The MIT score, also known as the Hsu score or Hsu-Zhang score, is a scoring method developed by Hsu (Feng Zhang's lab) based on data published in 2013 [5].
- Basis: Studied the indel mutation levels of more than 700 gRNA variants with 1-3 mismatches.
- Applications: MIT CRISPR design tool (replaced by CRISPick), CRISPOR.
③ Cutting Frequency Determination (CFD) :
- Reference: The score was referred to in Doench's 2016 paper [2]
- Basis: Based on the activity of 28,000 gRNAs with a single deletion/insertion/mutation.
- Scoring mode: A matrix for the scores of variations was generated. Since scores in the CFD matrix are less than 1, the more scores are multiplied, the lower the final score will become. The higher the score is, the higher off-target possibility there will be. Scores below 0.05 (some uses 0.023 as threshold) are considered as low off-target risk.
- Applications: Genscript, CRISPick.
Due to the importance of off-targeting in gene editing outcomes, especially in therapeutic development, implementing strategies to enhance specificity is crucial when high off-target is predicted.
Online Design Tool Examples
gRNAs are ranked based on a combination of their on-target and off-target scores and other related factors. There are multiple web-based gRNA design tools available, which uses different measurements for these parameters. Some of the most popular tools are listed below:
1. CRISPick (portals.broadinstitute.org): Developed by John Doench at the Broad Institute as one of the pioneering tools for gRNA design. It offers a simple interface and provides both on-target efficiency and off-target potential scores.
2. CHOPCHOP (chopchop.cbu.uib.no): CHOPCHOP is a versatile tool that supports various CRISPR-Cas systems beyond Cas9. It provides visual representations of potential off-target sites and allows for batch processing of multiple genes.
3. CRISPOR (crispor.tefor.net): CRISPOR provides a detailed off-target analysis with position-specific mismatch scoring. It also offers experimental considerations like restriction enzyme sites for cloning.
4. GenScript sgRNA Design Tool (www.genscript.com/tools/gRNA-design-tool): GenScript's sgRNA design tool utilizes Rule set 3 to assess on-target score and CFD to assess off-target score. These evaluation rules were selected for their updated logic derived from large scale experiments.
- Provides an overall score that balances on-target/off-target score, transcripts coverage, cutting positions (favoring sgRNAs closer to the 5' end of the CDS)
- Supports design for SpCas9 and AsCas12a (coming soon) with information of on-target/off-target scores using the latest logic, reasonable GC%, location, and target exons, as well as downstream ordering capability
- Displays gRNAs on the transcripts for easy selection according to different requests
- For knock-in experiments, an accompanying HDR Knock-In Design Tool is also available for HDR template & sgRNA design
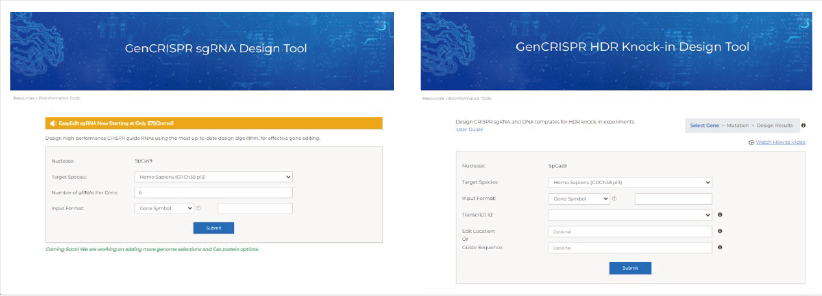
The simple and intuitive GenScript sgRNA and HDR Knock-in Design Tools
Designing sgRNAs for CRISPR-Cas9 genome editing is a complex process that requires careful consideration of various factors to achieve high on-target efficiency and specificity. We hope by leveraging these methods and resources, researchers can streamline their CRISPR experiments and accelerate their precision genome editing.
References:
[1] Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Doench JG, et.al. Nat Biotechnol. 2014
[2] Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Doench JG, et.al. Nat Biotechnol. 2016
[3] Accounting for small variations in the tracrRNA sequence improves sgRNA activity predictions for CRISPR screening. DeWeirdt PC, et.al. Nat Comm. 2022
[4] CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Moreno-Mateos MA, et.al. Nat Methods. 2015
[5] Massively parallel profiling and predictive modeling of the outcomes of CRISPR/Cas9-mediated double-strand break repair. Chen W, et. al. Nucleic Acids Res. 2019
[6] DNA targeting specificity of RNA-guided Cas9 nucleases. Hsu PD, et. al. Nat Biotechnol. 2013
Subscribe to have the latest weekly scientific insights delivery to your inbox!
* We'll never share your email address with a third-party.

-









































