| Gene Symbol | LOC542042 |
| Entrez Gene ID | 542042 |
| Full Name | MADS1 |
| Synonyms | GRMZM2G171365,SOC1,m5 |
| Gene Type | protein-coding |
| Organism | Zea mays |
-
REAGENT SERVICES
Hot!
-
Most Popular Services
-
Molecular Biology
-
Recombinant Antibody/Protein
-
Reagent Antibody
-
CRISPR Gene Editing
-
DNA Mutant Library
-
IVT RNA and LNP Formulations
-
Oligo Synthesis
-
Peptides
-
Cell Engineering
-
- Gene Synthesis FLASH Gene
- GenBrick™ Up to 200kb
- Gene Fragments Up to 3kb now
- Plasmid DNA Preparation Upgraded
- Cloning and Subcloning
- ORF cDNA Clones
- mRNA Plasmid Solutions New!
- Cell free mRNA Template New!
- AAV Plasmid Solutions New!
- Mutagenesis
- GenCircle™ Double-Stranded DNA New!
- GenSmart™ Online Tools
-
-
PRODUCTS
-
Most Popular Reagents
-
 Instruments
Instruments
-
Antibodies
-
ELISA Kits
-
Protein Electrophoresis and Blotting
-
Protein and Antibody Purification
-
Recombinant Proteins
-
Molecular Biology
-
Stable Cell Lines
-
Cell Isolation and Activation
-
 IVD Raw Materials
IVD Raw Materials
-
 Therapy Applications
Therapy Applications
-
Resources
-
- All Instruments
- Automated Protein and Antibody Purification SystemNew!
- Automated Plasmid MaxiprepHot!
- Automated Plasmid/Protein/Antibody Mini-scale Purification
- eBlot™ Protein Transfer System
- eStain™ Protein Staining System
- eZwest™ Lite Automated Western Blotting Device
- CytoSinct™ 1000 Cell Isolation Instrument
-
- Pharmacokinetics and Immunogenicity ELISA Kits
- Viral Titration QC ELISA Kits
- -- Lentivirus Titer p24 ELISA KitHot!
- -- MuLV Titer p30 ELISA KitNew!
- -- AAV2 and AAVX Titer Capsid ELISA Kits
- Residual Detection ELISA Kits
- -- T7 RNA Polymerase ELISA KitNew!
- -- BSA ELISA Kit, 2G
- -- Cas9 ELISA KitHot!
- -- Protein A ELISA KitHot!
- -- His tagged protein detection & purification
- dsRNA ELISA Kit
- Endonuclease ELISA Kit
- COVID-19 Detection cPass™ Technology Kits
-
- Automated Maxi-Plasmid PurificationHot!
- Automated Mini-Plasmid PurificationNew!
- PCR Reagents
- S.marcescens Nuclease Benz-Neburase™
- DNA Assembly GenBuilder™
- Cas9 / Cas12a / Cas13a Nucleases
- Base and Prime Editing Nucleases
- GMP Cas9 Nucleases
- CRISPR sgRNA Synthesis
- HDR Knock-in Template
- CRISPR Gene Editing Kits and Antibodies
-
![AmMag™ Quatro Automated Plasmid Purification]() AmMag™ Quatro automated plasmid purification
AmMag™ Quatro automated plasmid purification
-
![Anti-Camelid VHH]() MonoRab™ Anti-VHH Antibodies
MonoRab™ Anti-VHH Antibodies
-
![ELISA Kits]() ELISA Kits
ELISA Kits
-
![Precast Gels]() SurePAGE™ Precast Gels
SurePAGE™ Precast Gels
-
![Quatro ProAb Automated Protein and Antibody Purification System]() AmMag™ Quatro ProAb Automated Protein and Antibody Purification System
AmMag™ Quatro ProAb Automated Protein and Antibody Purification System
-
![Target Proteins]() Target Proteins
Target Proteins
-
![AmMag™ Quatro Automated Plasmid Purification]() AmMag™ Quatro automated plasmid purification
AmMag™ Quatro automated plasmid purification
-
![Stable Cell Lines]() Stable Cell Lines
Stable Cell Lines
-
![Cell Isolation and Activation]() Cell Isolation and Activation
Cell Isolation and Activation
-
 IVD Raw Materials
IVD Raw Materials
-
![Quick
Order]() Quick Order
Quick Order
-
![Quick
Order]() Quick Order
Quick Order
- APPLICATIONS
- RESOURCES
- ABOUT US
- SIGN IN My Account SIGN OUT
- REGISTER
ORF » Species Summary » Zea mays » LOC542042 cDNA ORF clone
LOC542042 cDNA ORF clone, Zea mays
-
Gene
-
Clones
gRNAs
-
Summary
-
mRNA and Protein(s)
mRNA Protein Name NM_001111682.1 NP_001105152.1 MADS1 -
Pathways from BioSystems

-
Gene Ontology

-
Bibliography
Related articles in PubMed
Flowering Time-Regulated Genes in Maize Include the Transcription Factor ZmMADS1.
Alter P, Bircheneder S, Zhou LZ, Schlüter U, Gahrtz M, Sonnewald U, Dresselhaus T
Plant physiology172(1)389-404(2016 09)The B73 maize genome: complexity, diversity, and dynamics.
Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, Minx P, Reily AD, Courtney L, Kruchowski SS, Tomlinson C, Strong C, Delehaunty K, Fronick C, Courtney B, Rock SM, Belter E, Du F, Kim K, Abbott RM, Cotton M, Levy A, Marchetto P, Ochoa K, Jackson SM, Gillam B, Chen W, Yan L, Higginbotham J, Cardenas M, Waligorski J, Applebaum E, Phelps L, Falcone J, Kanchi K, Thane T, Scimone A, Thane N, Henke J, Wang T, Ruppert J, Shah N, Rotter K, Hodges J, Ingenthron E, Cordes M, Kohlberg S, Sgro J, Delgado B, Mead K, Chinwalla A, Leonard S, Crouse K, Collura K, Kudrna D, Currie J, He R, Angelova A, Rajasekar S, Mueller T, Lomeli R, Scara G, Ko A, Delaney K, Wissotski M, Lopez G, Campos D, Braidotti M, Ashley E, Golser W, Kim H, Lee S, Lin J, Dujmic Z, Kim W, Talag J, Zuccolo A, Fan C, Sebastian A, Kramer M, Spiegel L, Nascimento L, Zutavern T, Miller B, Ambroise C, Muller S, Spooner W, Narechania A, Ren L, Wei S, Kumari S, Faga B, Levy MJ, McMahan L, Van Buren P, Vaughn MW, Ying K, Yeh CT, Emrich SJ, Jia Y, Kalyanaraman A, Hsia AP, Barbazuk WB, Baucom RS, Brutnell TP, Carpita NC, Chaparro C, Chia JM, Deragon JM, Estill JC, Fu Y, Jeddeloh JA, Han Y, Lee H, Li P, Lisch DR, Liu S, Liu Z, Nagel DH, McCann MC, SanMiguel P, Myers AM, Nettleton D, Nguyen J, Penning BW, Ponnala L, Schneider KL, Schwartz DC, Sharma A, Soderlund C, Springer NM, Sun Q, Wang H, Waterman M, Westerman R, Wolfgruber TK, Yang L, Yu Y, Zhang L, Zhou S, Zhu Q, Bennetzen JL, Dawe RK, Jiang J, Jiang N, Presting GG, Wessler SR, Aluru S, Martienssen RA, Clifton SW, McCombie WR, Wing RA, Wilson RK
Science (New York, N.Y.)326(5956)1112-5(2009 Nov)GRASSIUS: a platform for comparative regulatory genomics across the grasses.
Yilmaz A, Nishiyama MY Jr, Fuentes BG, Souza GM, Janies D, Gray J, Grotewold E
Plant physiology149(1)171-80(2009 Jan)The maize MADS box gene ZmMADS3 affects node number and spikelet development and is co-expressed with ZmMADS1 during flower development, in egg cells, and early embryogenesis.
Heuer S, Hansen S, Bantin J, Brettschneider R, Kranz E, Lörz H, Dresselhaus T
Plant physiology127(1)33-45(2001 Sep)GeneRIFs: Gene References Into Functions What's a GeneRIF?
The following LOC542042 gene cDNA ORF clone sequences were retrieved from the NCBI Reference Sequence Database (RefSeq). These sequences represent the protein coding region of the LOC542042 cDNA ORF which is encoded by the open reading frame (ORF) sequence. ORF sequences can be delivered in our standard vector, pcDNA3.1+/C-(K)DYK or the vector of your choice as an expression/transfection-ready ORF clone. Not the clone you want? Click here to find your clone.
-
Reference Sequences (Refseq)
CloneID OZa07680 Clone ID Related Accession (Same CDS sequence) NM_001111682.1 Accession Version NM_001111682.1 Latest version! Documents for ORF clone product in default vector Sequence Information ORF Nucleotide Sequence (Length: 699bp)
Protein sequence
SNPVector pcDNA3.1-C-(k)DYK or customized vector  User Manual
User ManualClone information Clone Map  MSDS
MSDSTag on pcDNA3.1+/C-(K)DYK C terminal DYKDDDDK tags ORF Insert Method CloneEZ™ Seamless cloning technology Insert Structure linear Update Date 1551628800000 Organism Zea mays Product MADS1 Comment Comment: PROVISIONAL REFSEQ: This record has not yet been subject to final NCBI review. The reference sequence was derived from AJ430642.1. ##Evidence-Data-START## Transcript exon combination :: AJ430642.1, BT055216.1 [ECO:0000332] RNAseq introns :: single sample supports all introns SAMN00780084, SAMN00780085 [ECO:0000348] ##Evidence-Data-END##
1
61
121
181
241
301
361
421
481
541
601
661ATGGTGCGGG GCAAGACGCA GATGAAGCGG ATAGAGAACC CGACCAGCCG CCAGGTCACC
TTCTCCAAGC GCCGCAACGG CCTGCTCAAG AAGGCGTTCG AGCTCTCCGT CCTCTGCGAC
GCCGAGGTCG CCCTCGTCGT CTTCTCCCCG CGCGGCAAGC TCTACGAATT CGCCAGCGGA
AGTGCGCAGA AAACGATTGA ACGTTATAGA ACATACACAA AGGATAATGT CAGCAACAAG
ACAGTGCAGC AGGATATTGA GCGAGTAAAA GCTGATGCGG ATGGCCTGTC AAAGAGACTC
GAAGCACTTG AAGCTTACAA AAGGAAACTT TTGGGTGAGA GGTTGGAAGA CTGCTCCATT
GAAGAGCTGC ACAGTTTGGA AGTCAAGCTT GAGAAGAGCC TGCATTGCAT CAGGGGAAGA
AAGACTGAGC TGCTGGAGGA GCAAGTCCGT AAGCTGAAGC AGAAGGAGAT GAGTCTGCGC
AAGAGCAACG AAGATTTGCG TGAAAAGTGC AAGAAGCAGC CGCCTGTGCC GATGGCTTCG
GCGCCGCCTC GTGCGCCGGC AGTCGACAAC GTGGAGGACG GTCACCGGGA GCCGAAGGAC
GACGGGATGG ACGTGGAGAC GGAGCTGTAC ATAGGATTGC CCGGCAGAGA CTACCGCTCA
AGCAAAGACA AGGCTGCAGT GGCGGTCAGG TCAGGCTAGThe stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
RefSeq NP_001105152.1 CDS 228..926 Translation 
Target ORF information:
RefSeq Version NM_001111682.1 Organism Zea mays Definition Zea mays MADS1 (LOC542042), mRNA. Target ORF information:
Epitope DYKDDDDK Bacterial selection AMPR Mammalian selection NeoR Vector pcDNA3.1+/C-(K)DYK 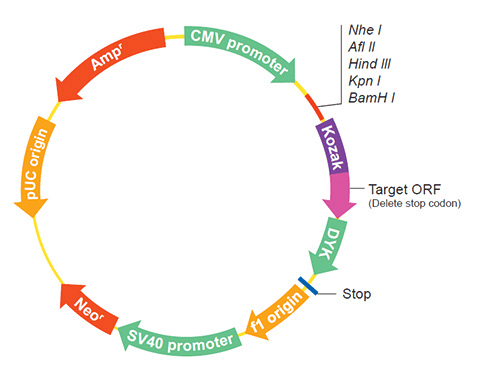 NM_001111682.1
NM_001111682.1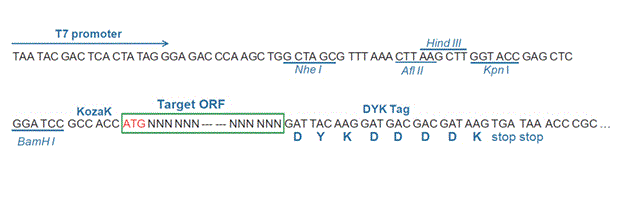
ORF Insert Sequence:
1
61
121
181
241
301
361
421
481
541
601
661ATGGTGCGGG GCAAGACGCA GATGAAGCGG ATAGAGAACC CGACCAGCCG CCAGGTCACC
TTCTCCAAGC GCCGCAACGG CCTGCTCAAG AAGGCGTTCG AGCTCTCCGT CCTCTGCGAC
GCCGAGGTCG CCCTCGTCGT CTTCTCCCCG CGCGGCAAGC TCTACGAATT CGCCAGCGGA
AGTGCGCAGA AAACGATTGA ACGTTATAGA ACATACACAA AGGATAATGT CAGCAACAAG
ACAGTGCAGC AGGATATTGA GCGAGTAAAA GCTGATGCGG ATGGCCTGTC AAAGAGACTC
GAAGCACTTG AAGCTTACAA AAGGAAACTT TTGGGTGAGA GGTTGGAAGA CTGCTCCATT
GAAGAGCTGC ACAGTTTGGA AGTCAAGCTT GAGAAGAGCC TGCATTGCAT CAGGGGAAGA
AAGACTGAGC TGCTGGAGGA GCAAGTCCGT AAGCTGAAGC AGAAGGAGAT GAGTCTGCGC
AAGAGCAACG AAGATTTGCG TGAAAAGTGC AAGAAGCAGC CGCCTGTGCC GATGGCTTCG
GCGCCGCCTC GTGCGCCGGC AGTCGACAAC GTGGAGGACG GTCACCGGGA GCCGAAGGAC
GACGGGATGG ACGTGGAGAC GGAGCTGTAC ATAGGATTGC CCGGCAGAGA CTACCGCTCA
AGCAAAGACA AGGCTGCAGT GGCGGTCAGG TCAGGCTAGThe stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
-
PubMed
Flowering Time-Regulated Genes in Maize Include the Transcription Factor ZmMADS1.
Plant physiology172(1)389-404(2016 09)
Alter P,Bircheneder S,Zhou LZ,Schlüter U,Gahrtz M,Sonnewald U,Dresselhaus T
The B73 maize genome: complexity, diversity, and dynamics.
Science (New York, N.Y.)326(5956)1112-5(2009 Nov)
Schnable PS,Ware D,Fulton RS,Stein JC,Wei F,Pasternak S,Liang C,Zhang J,Fulton L,Graves TA,Minx P,Reily AD,Courtney L,Kruchowski SS,Tomlinson C,Strong C,Delehaunty K,Fronick C,Courtney B,Rock SM,Belter E,Du F,Kim K,Abbott RM,Cotton M,Levy A,Marchetto P,Ochoa K,Jackson SM,Gillam B,Chen W,Yan L,Higginbotham J,Cardenas M,Waligorski J,Applebaum E,Phelps L,Falcone J,Kanchi K,Thane T,Scimone A,Thane N,Henke J,Wang T,Ruppert J,Shah N,Rotter K,Hodges J,Ingenthron E,Cordes M,Kohlberg S,Sgro J,Delgado B,Mead K,Chinwalla A,Leonard S,Crouse K,Collura K,Kudrna D,Currie J,He R,Angelova A,Rajasekar S,Mueller T,Lomeli R,Scara G,Ko A,Delaney K,Wissotski M,Lopez G,Campos D,Braidotti M,Ashley E,Golser W,Kim H,Lee S,Lin J,Dujmic Z,Kim W,Talag J,Zuccolo A,Fan C,Sebastian A,Kramer M,Spiegel L,Nascimento L,Zutavern T,Miller B,Ambroise C,Muller S,Spooner W,Narechania A,Ren L,Wei S,Kumari S,Faga B,Levy MJ,McMahan L,Van Buren P,Vaughn MW,Ying K,Yeh CT,Emrich SJ,Jia Y,Kalyanaraman A,Hsia AP,Barbazuk WB,Baucom RS,Brutnell TP,Carpita NC,Chaparro C,Chia JM,Deragon JM,Estill JC,Fu Y,Jeddeloh JA,Han Y,Lee H,Li P,Lisch DR,Liu S,Liu Z,Nagel DH,McCann MC,SanMiguel P,Myers AM,Nettleton D,Nguyen J,Penning BW,Ponnala L,Schneider KL,Schwartz DC,Sharma A,Soderlund C,Springer NM,Sun Q,Wang H,Waterman M,Westerman R,Wolfgruber TK,Yang L,Yu Y,Zhang L,Zhou S,Zhu Q,Bennetzen JL,Dawe RK,Jiang J,Jiang N,Presting GG,Wessler SR,Aluru S,Martienssen RA,Clifton SW,McCombie WR,Wing RA,Wilson RK
GRASSIUS: a platform for comparative regulatory genomics across the grasses.
Plant physiology149(1)171-80(2009 Jan)
Yilmaz A,Nishiyama MY Jr,Fuentes BG,Souza GM,Janies D,Gray J,Grotewold E
The maize MADS box gene ZmMADS3 affects node number and spikelet development and is co-expressed with ZmMADS1 during flower development, in egg cells, and early embryogenesis.
Plant physiology127(1)33-45(2001 Sep)
Heuer S,Hansen S,Bantin J,Brettschneider R,Kranz E,Lörz H,Dresselhaus T
-
-
Most Popular Services
-
Services & Products
-
News & Blogs
- “Wai Guo Ren” DNA encoded protein in living cells for the first time
- Gene Therapy for AADC: Targeting Neurons with AAV Vectors
- Therapeutic Antibodies Advancing Toward Approval for Clinical Use
- New Solutions Push the Limits of Engineered T Cells in Solid Tumors
- The Clinical Application of Neoantigens: A Promising Frontier in Cancer Immunotherapy
- Uncovering Immunogenic Peptides of Clinical Significance in CRISPR/Cas9
- CRISPR Screening to Optimize CAR Therapies | GenScript
- Cell and gene therapy at GenScript Biotech Global Forum | GenScript
-
-
-
PubMed
 Hi!Ask me about GenScript services and products! I can answer questions or connect you to a live person.
Hi!Ask me about GenScript services and products! I can answer questions or connect you to a live person. -




































