| Gene Symbol | TXNRD2 |
| Entrez Gene ID | 100455257 |
| Full Name | thioredoxin reductase 2 |
| Gene Type | protein-coding |
| Organism | Pongo abelii(Sumatran orangutan) |
-
REAGENT SERVICES
Hot!
-
Most Popular Services
-
Molecular Biology
-
Recombinant Antibody/Protein
-
Reagent Antibody
-
CRISPR Gene Editing
-
DNA Mutant Library
-
IVT RNA and LNP Formulations
-
Oligo Synthesis
-
Peptides
-
Cell Engineering
-
- Gene Synthesis FLASH Gene
- GenBrick™ Up to 200kb
- Gene Fragments Up to 3kb now
- Plasmid DNA Preparation Upgraded
- Cloning and Subcloning
- ORF cDNA Clones
- mRNA Plasmid Solutions New!
- Cell free mRNA Template New!
- AAV Plasmid Solutions New!
- Mutagenesis
- GenCircle™ Double-Stranded DNA New!
- GenSmart™ Online Tools
-
-
PRODUCTS
-
Most Popular Reagents
-
 Instruments
Instruments
-
Antibodies
-
ELISA Kits
-
Protein Electrophoresis and Blotting
-
Protein and Antibody Purification
-
Recombinant Proteins
-
Molecular Biology
-
Stable Cell Lines
-
Cell Isolation and Activation
-
 IVD Raw Materials
IVD Raw Materials
-
 Therapy Applications
Therapy Applications
-
Resources
-
- All Instruments
- Automated Protein and Antibody Purification SystemNew!
- Automated Plasmid MaxiprepHot!
- Automated Plasmid/Protein/Antibody Mini-scale Purification
- eBlot™ Protein Transfer System
- eStain™ Protein Staining System
- eZwest™ Lite Automated Western Blotting Device
- CytoSinct™ 1000 Cell Isolation Instrument
-
- Pharmacokinetics and Immunogenicity ELISA Kits
- Viral Titration QC ELISA Kits
- -- Lentivirus Titer p24 ELISA KitHot!
- -- MuLV Titer p30 ELISA KitNew!
- -- AAV2 and AAVX Titer Capsid ELISA Kits
- Residual Detection ELISA Kits
- -- T7 RNA Polymerase ELISA KitNew!
- -- BSA ELISA Kit, 2G
- -- Cas9 ELISA KitHot!
- -- Protein A ELISA KitHot!
- -- His tagged protein detection & purification
- dsRNA ELISA Kit
- Endonuclease ELISA Kit
- COVID-19 Detection cPass™ Technology Kits
-
- Automated Maxi-Plasmid PurificationHot!
- Automated Mini-Plasmid PurificationNew!
- PCR Reagents
- S.marcescens Nuclease Benz-Neburase™
- DNA Assembly GenBuilder™
- Cas9 / Cas12a / Cas13a Nucleases
- Base and Prime Editing Nucleases
- GMP Cas9 Nucleases
- CRISPR sgRNA Synthesis
- HDR Knock-in Template
- CRISPR Gene Editing Kits and Antibodies
-
![AmMag™ Quatro Automated Plasmid Purification]() AmMag™ Quatro automated plasmid purification
AmMag™ Quatro automated plasmid purification
-
![Anti-Camelid VHH]() MonoRab™ Anti-VHH Antibodies
MonoRab™ Anti-VHH Antibodies
-
![ELISA Kits]() ELISA Kits
ELISA Kits
-
![Precast Gels]() SurePAGE™ Precast Gels
SurePAGE™ Precast Gels
-
![Quatro ProAb Automated Protein and Antibody Purification System]() AmMag™ Quatro ProAb Automated Protein and Antibody Purification System
AmMag™ Quatro ProAb Automated Protein and Antibody Purification System
-
![Target Proteins]() Target Proteins
Target Proteins
-
![AmMag™ Quatro Automated Plasmid Purification]() AmMag™ Quatro automated plasmid purification
AmMag™ Quatro automated plasmid purification
-
![Stable Cell Lines]() Stable Cell Lines
Stable Cell Lines
-
![Cell Isolation and Activation]() Cell Isolation and Activation
Cell Isolation and Activation
-
 IVD Raw Materials
IVD Raw Materials
-
![Quick
Order]() Quick Order
Quick Order
-
![Quick
Order]() Quick Order
Quick Order
- APPLICATIONS
- RESOURCES
- ABOUT US
- SIGN IN My Account SIGN OUT
- REGISTER
ORF » Species Summary » Pongo abelii » TXNRD2 cDNA ORF clone
TXNRD2 cDNA ORF clone, Pongo abelii(Sumatran orangutan)
-
Gene
-
Clones
gRNAs
-
Summary
-
mRNA and Protein(s)
mRNA Protein Name NM_001256472.1 NP_001243401.1 thioredoxin reductase 2, mitochondrial -
Pathways from BioSystems

-
Gene Ontology

The following TXNRD2 gene cDNA ORF clone sequences were retrieved from the NCBI Reference Sequence Database (RefSeq). These sequences represent the protein coding region of the TXNRD2 cDNA ORF which is encoded by the open reading frame (ORF) sequence. ORF sequences can be delivered in our standard vector, pcDNA3.1+/C-(K)DYK or the vector of your choice as an expression/transfection-ready ORF clone. Not the clone you want? Click here to find your clone.
-
Reference Sequences (Refseq)
CloneID OSa22259 Clone ID Related Accession (Same CDS sequence) NM_001256472.1 Accession Version NM_001256472.1 Latest version! Documents for ORF clone product in default vector Sequence Information ORF Nucleotide Sequence (Length: 1575bp)
Protein sequence
SNPVector pcDNA3.1-C-(k)DYK or customized vector  User Manual
User ManualClone information Clone Map  MSDS
MSDSTag on pcDNA3.1+/C-(K)DYK C terminal DYKDDDDK tags ORF Insert Method CloneEZ™ Seamless cloning technology Insert Structure linear Update Date 1529942400000 Organism Pongo abelii(Sumatran orangutan) Product thioredoxin reductase 2, mitochondrial Comment Comment: VALIDATED REFSEQ: This record has undergone validation or preliminary review. The reference sequence was derived from NDHI03003715.1. On Feb 2, 2012 this sequence version replaced XM_002830854.1. Sequence Note: The RefSeq transcript and protein were derived from genomic sequence to make the sequence consistent with the reference genome assembly. The genomic coordinates used for the transcript record were based on alignments. ##Evidence-Data-START## RNAseq introns :: single sample supports all introns SAMEA2058381, SAMEA2058382 [ECO:0000348] ##Evidence-Data-END## ##RefSeq-Attributes-START## gene product(s) localized to mito. :: inferred from homology protein contains selenocysteine :: inferred from conservation ##RefSeq-Attributes-END## COMPLETENESS: complete on the 3' end.
1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561ATGGCGGCGA TGGCGGTGGC GCTGCGGGGA TTAGGAGGGC GCTTCCGGTG GCGGACGCAG
GCCGTGGCGG GCGGGGTGCG GGGCGCGGCG CGGGGCGCAG CAGCAGGTCA GCGGGACTAT
GATCTCCTGG TGGTCGGCGG GGGATCTGGC GGCTTGGCTT GTGCCAAGGA GGCCGCTCAG
CTGGGAAGGA AGGTGGCCGT GGTGGACTAC GTGGAACCTT CTCCCCAAGG CACCCGGTGG
GGCCTCGGCG GCACCTGCGT CAACGTGGGC TGCATCCCCA AGAAGCTGAT GCACCAGGCG
GCACTGCTGG GGGGACTGAT CCAAGATGCC CCCCACTATG GCTGGGAGGT GGCCCAGCCC
GTGCTGCATG ACTGGAGGAA GATGGCAGAA GCTGTTCAAA ATCACGTGAA ATCCTTGAAC
TGGGGCCACC GTGTCCAGCT TCAGGACAGA AAAGTCAAGT ACTTTAACAT CAAAGCCAGC
TTTGTTGACG AGCACACGGT TTGTGGCGTT GCCAAAGGTG GGAAAGAGAT TCTGCTATCA
GCTGACCACA TCATCATTGC TACTGGAGGG CGGCCGAGAT ACCCCACGCA CATCGAAGGT
GCCTTGGAAT ATGGAATCAC AAGTGATGAC ATCTTCTGGC TGAAGGAATC CCCTGGAAAA
ACGTTGGTGG TTGGGGCCAG CTATGTGGCC CTGGAGTGTG CTGGCTTCCT CACCGGGATT
GGGCTGGACA CCACCATCAT GATGCGCAGC ATCCCCCTCC GCGGCTTTGA CCAGCAAATG
TCCTCCTTGG TCATAGAGCA CATGGCATCT CATGGCACCC GGTTCCTGAG GGGCTGTGCC
CCCTCCCGGG TCAGGAGGCT CCCTGATGGC CAGCTGCAGG TCACCTGGGA GGACCGCGCC
ACCGGCAAGG AGGACATGGG CACCTTCGAC ACCGTCCTGT GGGCCATAGG TCGAGTCCCA
GACACCAGAA GTCTGAATTT GGAGAAGGCT GGGGTAGATA CTAGCCCCGA CACTCAGAAG
ATCCTGGTGG ACTCCCGGGA AGCCACCTCT GTGCCCCACA TCTACGCCAT TGGTGACGTG
GTGGAGGGGC GGCCTGAGCT GACACCCACA GCGATCATGG CCGGGAGGCT CCTGGTGCAG
CGGCTCTTCG GCGGGTCCTC AGATCTGATG GACTACGACA ATGTTCCCAC GACCGTCTTC
ACCCCGCTGG AGTATGGCTG TGTGGGGCTG TCCGAGGAGG AGGCAGTGGC TCGCCACGGG
CAGGAGCATG TTGAGGTCTA TCACGCCCAT TATAAACCAC TGGAGTTCAC GGTGGCTGGA
CGGGATGCAT CCCAGTGTTA TGTAAAGATG GTGTGCCTGA GGGAGCCCCC GCAGCTGCTG
CTGGGCCTGC ACTTCCTCGG CCCCAACGCA GGCGAAGTTA CTCAAGGATT TGCTCTGGGG
ATCAAGTGTG GGGCCTCCTA TGCGCAGGTG ATGCGGACCG TGGGTATCCA TCCCACATGC
TCTGAGGAGG TAGTCAAGCT GCGCATCTCC AAGCGCTCAG GCCTGGACCC CACGGTGACA
GGCTGCTGAG GGTAAThe stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
RefSeq NP_001243401.1 CDS 190..1764 Translation 
Target ORF information:
RefSeq Version NM_001256472.1 Organism Pongo abelii(Sumatran orangutan) Definition Pongo abelii thioredoxin reductase 2 (TXNRD2), mRNA. Target ORF information:
Epitope DYKDDDDK Bacterial selection AMPR Mammalian selection NeoR Vector pcDNA3.1+/C-(K)DYK 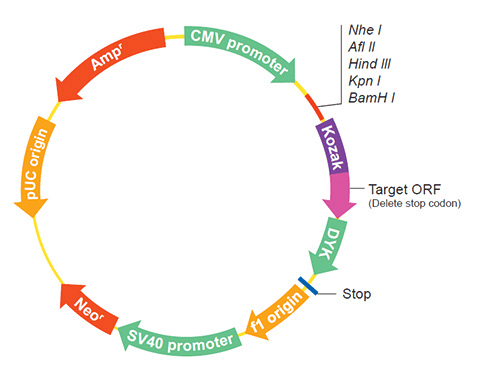 NM_001256472.1
NM_001256472.1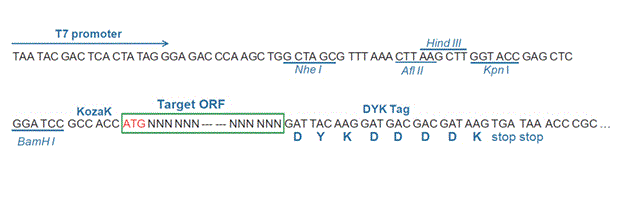
ORF Insert Sequence:
1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561ATGGCGGCGA TGGCGGTGGC GCTGCGGGGA TTAGGAGGGC GCTTCCGGTG GCGGACGCAG
GCCGTGGCGG GCGGGGTGCG GGGCGCGGCG CGGGGCGCAG CAGCAGGTCA GCGGGACTAT
GATCTCCTGG TGGTCGGCGG GGGATCTGGC GGCTTGGCTT GTGCCAAGGA GGCCGCTCAG
CTGGGAAGGA AGGTGGCCGT GGTGGACTAC GTGGAACCTT CTCCCCAAGG CACCCGGTGG
GGCCTCGGCG GCACCTGCGT CAACGTGGGC TGCATCCCCA AGAAGCTGAT GCACCAGGCG
GCACTGCTGG GGGGACTGAT CCAAGATGCC CCCCACTATG GCTGGGAGGT GGCCCAGCCC
GTGCTGCATG ACTGGAGGAA GATGGCAGAA GCTGTTCAAA ATCACGTGAA ATCCTTGAAC
TGGGGCCACC GTGTCCAGCT TCAGGACAGA AAAGTCAAGT ACTTTAACAT CAAAGCCAGC
TTTGTTGACG AGCACACGGT TTGTGGCGTT GCCAAAGGTG GGAAAGAGAT TCTGCTATCA
GCTGACCACA TCATCATTGC TACTGGAGGG CGGCCGAGAT ACCCCACGCA CATCGAAGGT
GCCTTGGAAT ATGGAATCAC AAGTGATGAC ATCTTCTGGC TGAAGGAATC CCCTGGAAAA
ACGTTGGTGG TTGGGGCCAG CTATGTGGCC CTGGAGTGTG CTGGCTTCCT CACCGGGATT
GGGCTGGACA CCACCATCAT GATGCGCAGC ATCCCCCTCC GCGGCTTTGA CCAGCAAATG
TCCTCCTTGG TCATAGAGCA CATGGCATCT CATGGCACCC GGTTCCTGAG GGGCTGTGCC
CCCTCCCGGG TCAGGAGGCT CCCTGATGGC CAGCTGCAGG TCACCTGGGA GGACCGCGCC
ACCGGCAAGG AGGACATGGG CACCTTCGAC ACCGTCCTGT GGGCCATAGG TCGAGTCCCA
GACACCAGAA GTCTGAATTT GGAGAAGGCT GGGGTAGATA CTAGCCCCGA CACTCAGAAG
ATCCTGGTGG ACTCCCGGGA AGCCACCTCT GTGCCCCACA TCTACGCCAT TGGTGACGTG
GTGGAGGGGC GGCCTGAGCT GACACCCACA GCGATCATGG CCGGGAGGCT CCTGGTGCAG
CGGCTCTTCG GCGGGTCCTC AGATCTGATG GACTACGACA ATGTTCCCAC GACCGTCTTC
ACCCCGCTGG AGTATGGCTG TGTGGGGCTG TCCGAGGAGG AGGCAGTGGC TCGCCACGGG
CAGGAGCATG TTGAGGTCTA TCACGCCCAT TATAAACCAC TGGAGTTCAC GGTGGCTGGA
CGGGATGCAT CCCAGTGTTA TGTAAAGATG GTGTGCCTGA GGGAGCCCCC GCAGCTGCTG
CTGGGCCTGC ACTTCCTCGG CCCCAACGCA GGCGAAGTTA CTCAAGGATT TGCTCTGGGG
ATCAAGTGTG GGGCCTCCTA TGCGCAGGTG ATGCGGACCG TGGGTATCCA TCCCACATGC
TCTGAGGAGG TAGTCAAGCT GCGCATCTCC AAGCGCTCAG GCCTGGACCC CACGGTGACA
GGCTGCTGAG GGTAAThe stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
-
PubMed
-
-
Most Popular Services
-
Services & Products
-
News & Blogs
- New Alzheimer's Biomarkers may Support Early Diagnosis
- CRISPR/Cas9 Used to Edit Gene Variance in Human Stem cell-derived β cells from a Patient with Monogenic Diabetes and Reverse Preexisting Diabetes in Mice
- The Clinical Application of Neoantigens: A Promising Frontier in Cancer Immunotherapy
- Multiplex Editing T Cells with BE and Cas12 | GenScript
- Using Next Generation Sequencing, Scientist have Discovered an Answer to a 4,000 Year Old Mummy Mystery
- A New Strategy Evades Bacterial Resistance-Targeting Beta-Lactamases with Synthetic Peptides to Improve Antibiotic Performance
- Discovering Antibodies with Broader Epitope Specificities
- NGS - A Tool to Identification Variants in Acute Myeloid Leukemia Patients
-
-
-
PubMed
 Hi!Ask me about GenScript services and products! I can answer questions or connect you to a live person.
Hi!Ask me about GenScript services and products! I can answer questions or connect you to a live person. -




































