Escherichia coli: (Genetic code: Standard)
| Triplet | Amino acid | Fraction | Frequency/ Thousand | Number | Triplet | Amino acid | Fraction | Frequency/ Thousand | Number |
|---|---|---|---|---|---|---|---|---|---|
| TTT TTC TTA TTG |
F F L L |
0.58 0.42 0.14 0.13 |
22.1 16.0 14.3 13.0 |
80995 58774 52382 47500 |
TCT TCC TCA TCG |
S S S S |
0.17 0.15 0.14 0.14 |
10.4 9.1 8.9 8.5 |
38027 33430 32715 31146 |
| TAT TAC TAA TAG |
Y Y * * |
0.59 0.41 0.61 0.09 |
17.5 12.2 2.0 0.3 |
63937 44631 7356 989 |
TGT TGC TGA TGG |
C C * W |
0.46 0.54 0.30 1.00 |
5.2 6.1 1.0 13.9 |
19138 22188 3623 50991 |
| CTT CTC CTA CTG |
L L L L |
0.12 0.10 0.04 0.47 |
11.9 10.2 4.2 48.4 |
43449 37347 15409 177210 |
CCT CCC CCA CCG |
P P P P |
0.18 0.13 0.20 0.49 |
7.5 5.4 8.6 20.9 |
27340 19666 31534 76644 |
| CAT CAC CAA CAG |
H H Q Q |
0.57 0.43 0.34 0.66 |
12.5 9.3 14.6 28.4 |
45879 34078 53394 104171 |
CGT CGC CGA CGG |
R R R R |
0.36 0.36 0.07 0.11 |
20.0 19.7 3.8 5.9 |
73197 72212 13844 21552 |
| ATT ATC ATA ATG |
I I I M |
0.49 0.39 0.11 1.00 |
29.8 23.7 6.8 26.4 |
109072 86796 24984 96695 |
ACT ACC ACA ACG |
T T T T |
0.19 0.40 0.17 0.25 |
10.3 22.0 9.3 13.7 |
37842 80547 33910 50269 |
| AAT AAC AAA AAG |
N N K K |
0.49 0.51 0.74 0.26 |
20.6 21.4 35.3 12.4 |
75436 78443 129137 45459 |
AGT AGC AGA AGG |
S S R R |
0.16 0.25 0.07 0.04 |
9.9 15.2 3.6 2.1 |
36097 55551 13152 7607 |
| GTT GTC GTA GTG |
V V V V |
0.28 0.20 0.17 0.35 |
19.8 14.3 11.6 24.4 |
72584 52439 42420 89265 |
GCT GCC GCA GCG |
A A A A |
0.18 0.26 0.23 0.33 |
17.1 24.2 21.2 30.1 |
62479 88721 77547 110308 |
| GAT GAC GAA GAG |
D D E E |
0.63 0.37 0.68 0.32 |
32.7 19.2 39.1 18.7 |
119939 70394 143353 68609 |
GGT GGC GGA GGG |
G G G G |
0.35 0.37 0.13 0.15 |
25.5 27.1 9.5 11.3 |
93325 99390 34799 41277 |
Codon Usage is only one of many DNA sequence features that influence protein expression levels
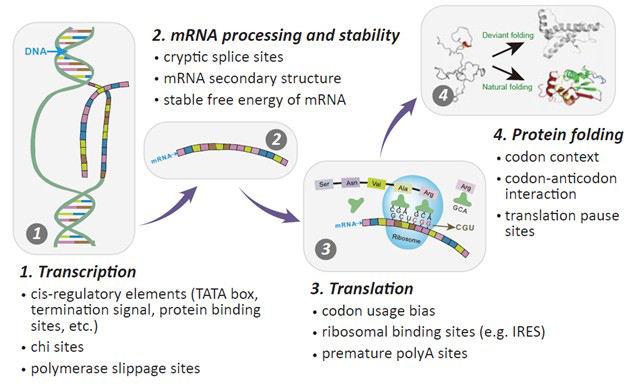
Can’t improve protein expression even after codon bias adjust?
A lots of parameters affect the protein expression besides codon bias:
- GC content
- CpG dinucleotides content
- Cryptic splicing sites
- Codon-context
- Negative CpG islands
- Premature PolyA sites
- Terminal signal
- TATA boxes
- mRNA secondary structure
- SD sequence
- RNA instability motif (ARE)
- RNA secondary structures
- Interaction of codon and anti-codon
- Internal chi sites and ribosomal binding sites
- Stable free energy of mRNA
GenScript OptimumGene™ algorithm provides a comprehensive solution strategy on optimizing all parameters that are critical to protein expression levels. It can significate increase protein expression level up to 50-fold, provided that the protein expression and purification methods are appropriately applied.







































