-
REAGENT SERVICES
Hot!
-
Most Popular Services
-
Molecular Biology
-
Recombinant Antibody/Protein
-
Reagent Antibody
-
CRISPR Gene Editing
-
DNA Mutant Library
-
IVT RNA and LNP Formulations
-
Oligo Synthesis
-
Peptides
-
Cell Engineering
-
- Gene Synthesis FLASH Gene
- GenBrick™ Up to 200kb
- Gene Fragments Up to 3kb now
- Plasmid DNA Preparation Upgraded
- Cloning and Subcloning
- ORF cDNA Clones
- mRNA Plasmid Solutions New!
- Cell free mRNA Template New!
- AAV Plasmid Solutions New!
- Mutagenesis
- GenCircle™ Double-Stranded DNA New!
- GenSmart™ Online Tools
-
-
PRODUCTS
-
Most Popular Reagents
-
 Instruments
Instruments
-
Antibodies
-
ELISA Kits
-
Protein Electrophoresis and Blotting
-
Protein and Antibody Purification
-
Recombinant Proteins
-
Molecular Biology
-
Stable Cell Lines
-
Cell Isolation and Activation
-
 IVD Raw Materials
IVD Raw Materials
-
 Therapy Applications
Therapy Applications
-
Resources
-
- All Instruments
- Automated Protein and Antibody Purification SystemNew!
- Automated Plasmid MaxiprepHot!
- Automated Plasmid/Protein/Antibody Mini-scale Purification
- eBlot™ Protein Transfer System
- eStain™ Protein Staining System
- eZwest™ Lite Automated Western Blotting Device
- CytoSinct™ 1000 Cell Isolation Instrument
-
- Pharmacokinetics and Immunogenicity ELISA Kits
- Viral Titration QC ELISA Kits
- -- Lentivirus Titer p24 ELISA KitHot!
- -- MuLV Titer p30 ELISA KitNew!
- -- AAV2 and AAVX Titer Capsid ELISA Kits
- Residual Detection ELISA Kits
- -- T7 RNA Polymerase ELISA KitNew!
- -- BSA ELISA Kit, 2G
- -- Cas9 ELISA KitHot!
- -- Protein A ELISA KitHot!
- -- His tagged protein detection & purification
- dsRNA ELISA Kit
- Endonuclease ELISA Kit
- COVID-19 Detection cPass™ Technology Kits
-
- Automated Maxi-Plasmid PurificationHot!
- Automated Mini-Plasmid PurificationNew!
- PCR Reagents
- S.marcescens Nuclease Benz-Neburase™
- DNA Assembly GenBuilder™
- Cas9 / Cas12a / Cas13a Nucleases
- Base and Prime Editing Nucleases
- GMP Cas9 Nucleases
- CRISPR sgRNA Synthesis
- HDR Knock-in Template
- CRISPR Gene Editing Kits and Antibodies
-
![AmMag™ Quatro Automated Plasmid Purification]() AmMag™ Quatro automated plasmid purification
AmMag™ Quatro automated plasmid purification
-
![Anti-Camelid VHH]() MonoRab™ Anti-VHH Antibodies
MonoRab™ Anti-VHH Antibodies
-
![ELISA Kits]() ELISA Kits
ELISA Kits
-
![Precast Gels]() SurePAGE™ Precast Gels
SurePAGE™ Precast Gels
-
![Quatro ProAb Automated Protein and Antibody Purification System]() AmMag™ Quatro ProAb Automated Protein and Antibody Purification System
AmMag™ Quatro ProAb Automated Protein and Antibody Purification System
-
![Target Proteins]() Target Proteins
Target Proteins
-
![AmMag™ Quatro Automated Plasmid Purification]() AmMag™ Quatro automated plasmid purification
AmMag™ Quatro automated plasmid purification
-
![Stable Cell Lines]() Stable Cell Lines
Stable Cell Lines
-
![Cell Isolation and Activation]() Cell Isolation and Activation
Cell Isolation and Activation
-
 IVD Raw Materials
IVD Raw Materials
-
![Quick
Order]() Quick Order
Quick Order
-
![Quick
Order]() Quick Order
Quick Order
- APPLICATIONS
- RESOURCES
- ABOUT US
- SIGN IN My Account SIGN OUT
- REGISTER
News & Blogs » Peptide News » New High-throughput Platform to Expedite Shared Neoantigen-Based Cancer Immunotherapies
New High-throughput Platform to Expedite Shared Neoantigen-Based Cancer Immunotherapies
Neoantigen-based immunotherapies, such as cancer vaccines, have proved efficient against melanoma. Reports from two recent trials found that an mRNA and a dendritic cell neoantigen vaccine could improve patient survival and induce long-lasting tumor-specific T-cell memory responses, respectively (Liu et al. 2022). Neoantigen-based therapies target novel antigens formed within tumors due to somatic mutations. Because these novel antigens are foreign, neoantigen-specific immunotherapies such as vaccines or T-cell transfer can target and kill cancer cells.
For neoantigens to have clinical value in these approaches, they must be sufficiently and ideally exclusively expressed at the tumor site, presented as part of a peptide-MHC complex, and able to stimulate an effective T cell response against tumor cells. However, identifying neoantigens of clinical utility is a challenging process, requiring multiple approaches such as next-generation sequencing to identify often thousands of mutations in resected tumor tissue, algorithms to predict neoantigen presentation, immunopeptidomics to confirm neoantigen expression and MHC presentation in the tumor, and functional assays to demonstrate immunogenicity or T cell reactivity (Becker and Riemer 2022). Therefore, the time required for neoantigen discovery represents a significant factor limiting prompt access to cancer immunotherapies at the clinic.

Neoantigen Discovery Workflow. “In typical neoantigen identification workflows, candidate tumor antigens are defined from next generation sequencing data (left side). Subsequent in silico binding predictions and in vitro immunogenicity tests identify potential tumor neoepitopes, which, however, are not necessarily presented on the tumor cells themselves. If the target epitope is not present, CD8+ T cells cannot identify the transformed cells and the tumor proliferates (middle). In contrast, mass spectrometry-based immunopeptidomics identifies truly presented and actionable epitopes. Immunotherapies targeting these epitopes lead to an immune attack by CD8+ T cells and tumor destruction (right side).” Created with BioRender.com. Retrieved from Becker and Riemer 2022, https://creativecommons.org/licenses/by/4.0/.
To circumvent this obstacle, immunotherapies can target shared neoantigens rather than unique or private tumor antigens. Shared neoantigens result from frequent mutations among cancer types and/or patients (Zhang et al. 2021). While the initial discovery and validation process for shared and private neoantigens is the same, once identified as immunogenic targets, shared neoantigens can help expedite therapies to patients harboring the same mutations.
Recent work published by scientists at Genentech aims to expand our knowledge base of shared neoantigens to provide more opportunities for off-the-shelf cancer immunotherapies. To this end, they have developed a new platform to identify shared neoepitope-MHC pairs.
The team focused on prevalent cancer point mutations and frequently expressed MHC alleles, which were evaluated to identify peptide-MHC binding pairs through a Time-Resolved Fluorescence Energy Transfer (TR-FRET) assay. The assay leveraged single MHC molecules preloaded with a UV-sensitive peptide and conjugated to a FRET acceptor (shown below). Following cleavage of the pre-loaded peptide, MHCs remained stable and capable of FRET signaling only upon binding to a matching neoepitope. Therefore, by monitoring the TR-FRET signals, Gurung and colleagues could identify stable neoepitope-MHC pairs.

TR-FRET assay to identify shared neoepitope-MHC pairs. “Schematic diagram of the TR-FRET assay used to measure stable neoepitope–HLA binding. In brief, HLA monomers bound to UV-cleavable peptides are exposed to UV light in the presence of mutation-bearing candidate neoepitopes. Successful exchange of the candidate peptide will lead to complex stabilization and TR-FRET emission (top). Unsuccessful exchange will lead to aggregation and no TR-FRET emission (bottom).” Retrieved from Gurung et al. 2023. Only panel b of Figure 1 and legend are shown https://creativecommons.org/licenses/by/4.0/
Gurung et al. found agreement between some of the neoepitope-MHC pairs identified experimentally with those predicted computationally, yet each approach also identified new pairs. To verify the identified pairs in the context of cellular processing, the team leveraged MHC monoallelic cell lines transfected with multiple concatenated neoantigen sequences. Thereby, neoepitope-MHC binding was verified by immunopeptidomics through targeted and untargeted mass spectral (MS) approaches. More than 80 neoepitope-MHC allele pairs were identified, with over half corresponding to novel pairs. Significantly, the identified pairs agreed with their initial computational and TR-FRET findings, demonstrating the strengths of their discovery workflow.

Immunopeptidomics Workflow. Mass spectral identification of neoantigen-MHC pairs using engineered MHC monoallelic cell lines transfected with a cassette of 47 different neoantigens peptides. Retrieved from Gurung et al. 2023 Figure 3, only panel a is shown. https://creativecommons.org/licenses/by/4.0/
Lastly, the ultimate validation of neoantigen peptide-MHC pairs as targets of clinical value depends on their ability to induce productive T cell responses. Moreover, identifying the reactive T cell populations and their respective receptor sequences expands opportunities to develop ready-made immunotherapies. Therefore, to this end, the team leveraged synthetic peptide pools corresponding to the identified neoantigens to validate their capacity to activate patient-derived CD8+ T cells. Next, the T cell receptors (TCR) of neoepitope-responsive cells were expressed in primary T cells for activity assays. Through this approach, Gurung et al. identified individual neoepitope-MHC pairs with the capacity to interact with specific TCRs to effect CD8+ T cell-mediated cytotoxic killing.
For TCR discovery, “A total of 376 predicted and MS-identified neoantigen-derived peptides were synthesized (GenScript).” Gurung et al. 2023
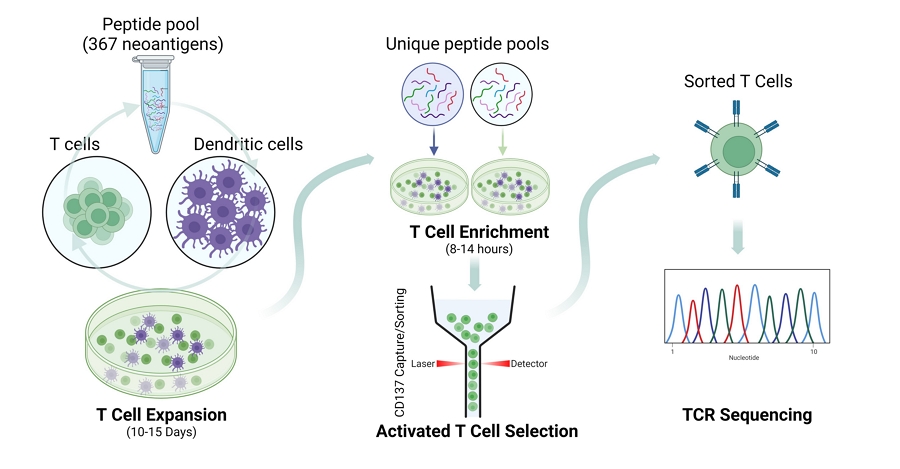
Validating neoepitope-T Cell Reactivity with Synthetic Peptides. Briefly, donor derived CD8+ T cells were first expanded by incubation with an anti-CD3 antibody. Alternatively, expansion was achieved by co-culture of T cells with autologous dendritic cells pulsed with a pool of 376 synthetic peptides, corresponding to all identified neoepitopes. After this initial expansion, CD8+ T cells were again co-cultured with peptide pools having unique neoepitope combinations. Following enrichment, activated T cells were sorted for TCR sequencing analysis. Created with BioRender.
Overall, Gurung and colleagues have successfully developed a pipeline that leverages various high-throughput and functional assays with the capacity to identify novel neoepitope-MHC pairs that could serve as effective cancer immunotherapy targets for a broader patient population.
GenQuiz: New High-throughput Platform to Expedite Shared Neoantigen-Based Cancer Immunotherapies
Reference
-
Becker, J. P., & Riemer, A. B. (2022). The Importance of Being Presented: Target Validation by Immunopeptidomics for Epitope-Specific Immunotherapies. Frontiers in Immunology, 13, 883989. https://doi.org/10.3389/FIMMU.2022.883989/BIBTEX
-
Gurung, H. R., Heidersbach, A. J., Darwish, M., Chan, P. P. F., Li, J., Beresini, M., Zill, O. A., Wallace, A., Tong, A. J., Hascall, D., Torres, E., Chang, A., Lou, K. ‘Hei W., Abdolazimi, Y., Hammer, C., Xavier-Magalhães, A., Marcu, A., Vaidya, S., Le, D. D., … Rose, C. M. (2023). Systematic discovery of neoepitope–HLA pairs for neoantigens shared among patients and tumor types. Nature Biotechnology 2023, 1–11. https://doi.org/10.1038/s41587-023-01945-y
-
Liu, J., Fu, M., Wang, M., Wan, D., Wei, Y., & Wei, X. (2022). Cancer vaccines as promising immuno-therapeutics: platforms and current progress. Journal of Hematology & Oncology 2022 15:1, 15(1), 1–26. https://doi.org/10.1186/S13045-022-01247-X
-
Zhang, Z., Lu, M., Qin, Y., Gao, W., Tao, L., Su, W., & Zhong, J. (2021). Neoantigen: A New Breakthrough in Tumor Immunotherapy. Frontiers in Immunology, 12, 672356. https://doi.org/10.3389/FIMMU.2021.672356/BIBTEX
Subscribe to Receive Updates
& Promotions From GenScript* We'll never share your email address with a third-party.
Latest News & Blogs
Find More Peptide News
The Future of Neoantigen Vaccines: How Nanoparticles, Liposomes, and Viral Vectors Are Revolutionizing Treatment as Novel delivery System
Sep 22, 2025

Unveiling the Potential of GLP-1 Receptor Agonists and Multi-Target Therapies
Jul 07, 2025

Challenges in Neoantigen Peptide Manufacturing | GenScript
Apr 28, 2025

Challenges & Innovations in Neoantigen Peptide Vaccine Production
Apr 02, 2025

The Clinical Application of Neoantigens: A Promising Frontier in Cancer Immunotherapy
Feb 25, 2025

Delving into the World of Cyclic Peptides: Harnessing the Power of Circular Molecules for Innovative Therapeutic Solutions
May 14, 2024

-






































