-
REAGENT SERVICES
Hot!
-
Most Popular Services
-
Molecular Biology
-
Recombinant Antibody/Protein
-
Reagent Antibody
-
CRISPR Gene Editing
-
DNA Mutant Library
-
IVT RNA and LNP Formulations
-
Oligo Synthesis
-
Peptides
-
Cell Engineering
-
- Gene Synthesis FLASH Gene
- GenBrick™ Up to 200kb
- Gene Fragments Up to 3kb now
- Plasmid DNA Preparation Upgraded
- Cloning and Subcloning
- ORF cDNA Clones
- mRNA Plasmid Solutions New!
- Cell free mRNA Template New!
- AAV Plasmid Solutions New!
- Mutagenesis
- GenCircle™ Double-Stranded DNA New!
- GenSmart™ Online Tools
-
-
PRODUCTS
-
Most Popular Reagents
-
 Instruments
Instruments
-
Antibodies
-
ELISA Kits
-
Protein Electrophoresis and Blotting
-
Protein and Antibody Purification
-
Recombinant Proteins
-
Molecular Biology
-
Stable Cell Lines
-
Cell Isolation and Activation
-
 IVD Raw Materials
IVD Raw Materials
-
 Therapy Applications
Therapy Applications
-
Resources
-
- All Instruments
- Automated Protein and Antibody Purification SystemNew!
- Automated Plasmid MaxiprepHot!
- Automated Plasmid/Protein/Antibody Mini-scale Purification
- eBlot™ Protein Transfer System
- eStain™ Protein Staining System
- eZwest™ Lite Automated Western Blotting Device
- CytoSinct™ 1000 Cell Isolation Instrument
-
- Pharmacokinetics and Immunogenicity ELISA Kits
- Viral Titration QC ELISA Kits
- -- Lentivirus Titer p24 ELISA KitHot!
- -- MuLV Titer p30 ELISA KitNew!
- -- AAV2 and AAVX Titer Capsid ELISA Kits
- Residual Detection ELISA Kits
- -- T7 RNA Polymerase ELISA KitNew!
- -- BSA ELISA Kit, 2G
- -- Cas9 ELISA KitHot!
- -- Protein A ELISA KitHot!
- -- His tagged protein detection & purification
- dsRNA ELISA Kit
- Endonuclease ELISA Kit
- COVID-19 Detection cPass™ Technology Kits
-
- Automated Maxi-Plasmid PurificationHot!
- Automated Mini-Plasmid PurificationNew!
- PCR Reagents
- S.marcescens Nuclease Benz-Neburase™
- DNA Assembly GenBuilder™
- Cas9 / Cas12a / Cas13a Nucleases
- Base and Prime Editing Nucleases
- GMP Cas9 Nucleases
- CRISPR sgRNA Synthesis
- HDR Knock-in Template
- CRISPR Gene Editing Kits and Antibodies
-
![AmMag™ Quatro Automated Plasmid Purification]() AmMag™ Quatro automated plasmid purification
AmMag™ Quatro automated plasmid purification
-
![Anti-Camelid VHH]() MonoRab™ Anti-VHH Antibodies
MonoRab™ Anti-VHH Antibodies
-
![ELISA Kits]() ELISA Kits
ELISA Kits
-
![Precast Gels]() SurePAGE™ Precast Gels
SurePAGE™ Precast Gels
-
![Quatro ProAb Automated Protein and Antibody Purification System]() AmMag™ Quatro ProAb Automated Protein and Antibody Purification System
AmMag™ Quatro ProAb Automated Protein and Antibody Purification System
-
![Target Proteins]() Target Proteins
Target Proteins
-
![AmMag™ Quatro Automated Plasmid Purification]() AmMag™ Quatro automated plasmid purification
AmMag™ Quatro automated plasmid purification
-
![Stable Cell Lines]() Stable Cell Lines
Stable Cell Lines
-
![Cell Isolation and Activation]() Cell Isolation and Activation
Cell Isolation and Activation
-
 IVD Raw Materials
IVD Raw Materials
-
![Quick
Order]() Quick Order
Quick Order
-
![Quick
Order]() Quick Order
Quick Order
- APPLICATIONS
- RESOURCES
- ABOUT US
- SIGN IN My Account SIGN OUT
- REGISTER

![Case Study: Representative High-Throughput DNA Library Assembly Case Study: Representative High-Throughput DNA Library Assembly]()
High-Throughput DNA Library Assembly Services
GenScript Builds Your Design Faster!
Home » High-Throughput DNA Library Assembly » Case Study - Representative Combinatorial DNA Library
Overview
Our client constructed a four-gene metabolic pathway in yeast to convert a substrate to the precursor of a small molecule drug. The idea is simple, they put the four different pathway genes under the control of four constitutive promoters, then put the whole pathway in a low copy number plasmid in a yeast strain. However, they are not satisfied with titer of the resulting yeast strain, as it is far from sufficient to reach the commercial production level. To optimize the pathway and improve the efficiency, they need to optimize the enzymes for each of the reaction.
 Figure 1. Using yeast cells to produce the precursor for a small molecule drug.
Figure 1. Using yeast cells to produce the precursor for a small molecule drug.Client requested the construction of a four-gene metabolic pathway with four constitutive promoters in a low-copy number vector. Fifty variants for each pathway gene were identified and designed through database blasting and literature search. These gene variants were randomly inserted into each of the coding DNA sequence (CDS) positions in the pathway, making the theoretical library size 6,250,000.
 Figure 2. Workflow for combinatorial pathway assembly and screening.
Figure 2. Workflow for combinatorial pathway assembly and screening.Library was constructed as a pool by putting all fragments into the reaction. Then, 2,000 colonies were randomly picked and tested for positive inserts. More than 80% positive insertion rate was achieved. In addition, all plasmids with inserts were mini-prepped with high-throughput platform, followed by the transformation of plasmids into yeast cells for performance testing and screening.
 Figure 3. 100% library diversity confirmed by sequencing to ensure maximum coverage.
Figure 3. 100% library diversity confirmed by sequencing to ensure maximum coverage.48 randomly-picked positive clones were analyzed by sequencing. Results demonstrated 100% diversity and little assembly bias.
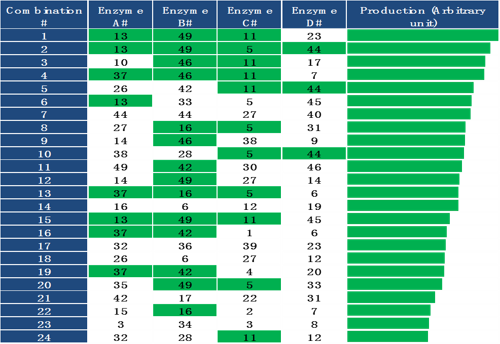 Figure 4. Isolation and analysis of clones with improved performance.
Figure 4. Isolation and analysis of clones with improved performance.After screening all positive clones for their performance, the top 96 best performing clones were sequenced. The enrichments of some variants were higher than others in the good-performing clones. These good-performing enzyme variants can be further combinatorically tested in the next round of optimization.
Mutagenesis Expression | Recombinant Mutagenesis & rAb Services - GenScript protein2022Get in Touch
with GenScript Custom
Mutagenesis Services
 Hi!Ask me about GenScript services and products! I can answer questions or connect you to a live person.
Hi!Ask me about GenScript services and products! I can answer questions or connect you to a live person. -





































