

Celebrating 18 years of CRISPR
With the BEST CRISPR cell line offer on the market!
Starting at only $7,000/KO Cell Line!
GenScript licensed CRISPR/Cas9 technology from the Broad Institute of MIT and Harvard, and provides full CRISPR-based gene editing service to produce genetically modified cells lines using any mammalian cell line and targeting any gene.
* It is preferred that customers provide their own cell lines, despite available cell lines can be purchased by GenScript for an additional fee. Please note that we currently do not provide genome editing service for primary cells, stem cells or iPS cells.
** Validation services for knock-out cell lines can be found under the "Add On Services" tab.
CRISPR workflow
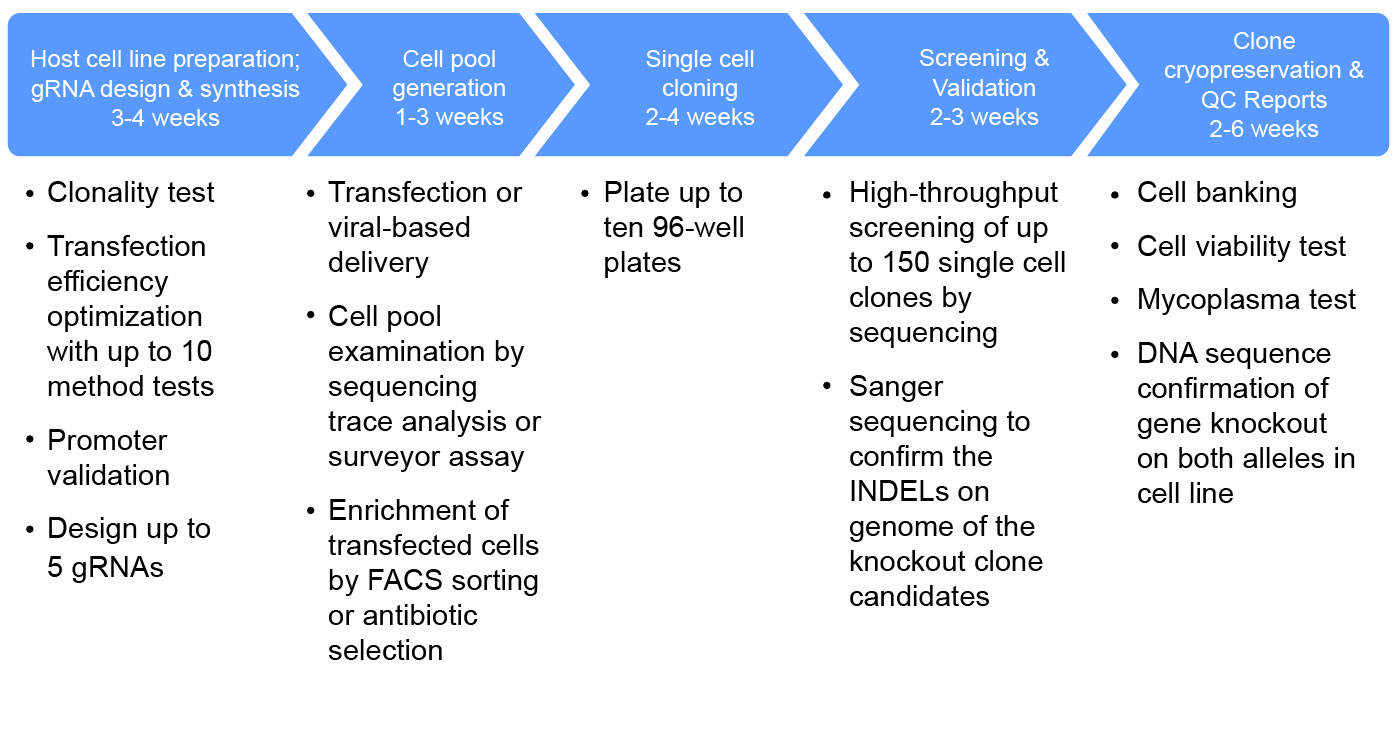
Optional Services
| Add-On Service | Description |
|---|---|
| Reverse transcription (RT)-PCR | Validate the knockout clones to carry the INDELs on CDS at mRNA level by sequencing the RT-PCR product. |
| Western blot | Validate the knockout clones by western blot. A validated antibody in the host cells with specific binding to target protein (RNAi or knockout) must be provided. |
| FACS analysis | Validate the knockout clones by FACS analysis. A validated antibody in the host cells with specific binding to target protein (RNAi or knockout) must be provided. |
| Off-target analysis | Characterize one knockout clone by sequencing the top 10 of potential off-target sites. |
| One additional clone | Sequence additional single clones to identify one additional knockout clone. QC and deliver one additional knockout clone. |
Related services
- gRNA sequence databases Validated gRNA sequences for efficient, specific targeting of WTCas9 or SAM.
- CRISPR gRNA constructs: all-in-one and dual WT SpCas9, SaCas9 and Nickase plasmids for single gRNA expression.
- SAM constructs for transcription activation.
- CRISPR gRNA libraries: genome-wide knock-out ( GeCKO v2), transcription activation ( SAM) and pathway-focused libraries for efficient screening.
- Microbial genome editing service using a novel λ Red – CRISPR/Cas system.
- GenScript offers lentiviral based service, CellPower™ and other cell line services.
- GenCRISPR™/Cas9 genome editing products-purified Cas9 proteins and related products to facilitate high efficient genome engineering.
Case Study I
- K-ras locus in human colon cancer cell line, HCT116, was knocked-out by creating indels in exon 4, as shown in the targeting strategy (Fig. 1).
- Viral encapsulation of Cas9 and gRNA delivery particles were carried out, viral titer was optimized for HCT116 cells.
- Individual clones were Sanger sequenced to select for homozygous knock-out indels of K-ras at exon 4. Sanger sequencing data (Fig. 2).
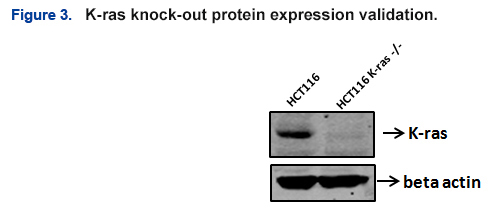
- Absence of K-ras expression in the selected clone was confirmed via Western Blot (Fig. 3).


Case Study II
- A sequence optimized gRNA was designed and synthesized to target a specific region on the GS allele. DG44 cells were transfected with the construct and the cell pool was analyzed by Sanger sequencing.
- Several single clones were obtained and Sanger sequence analyzed. A single clone containing a frame shift mutation was carried forward (Fig. 1).
- The GS knockout clone was also analyzed for GS protein expression (Fig. 2).
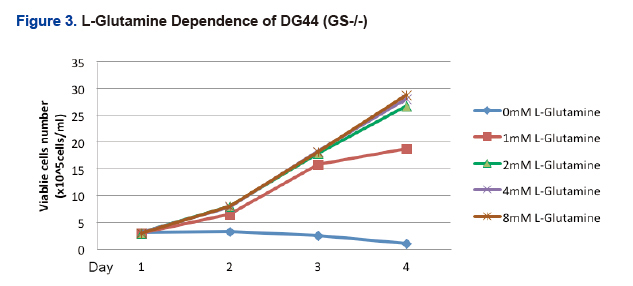
- To assess loss of function, GS knockouts were grown in the presence or absence of increasing concentrations of glutamine (Fig. 3). GS knockouts were unable to grow in the absence of glutamine. However growth improved in the presence of increasing concentrations of glutamine, thereby indicating a functional loss in the GS knockout cell line.
- In conclusion, successful targeting of GS generated a single clone that was fully validated for lack of expression and GS function.


Case Study III
- gRNA pairs are designed to target the flanking locus of a full gene to delete a full gene on genome. gRNAs pairs are optimized with highest cleavage efficiency in the Jurkat cells by GenCRISPR™ technology (Figure A).
- The paired gRNAs are co-transfected to the Jurkat cells and enriched by FACS sorting. The deletion efficiency is evaluated by PCR. It is 21% in the transfected cell pool as calculated from light intensity of band (Figure B).
- 200 single clones are screened with PCR in a high-throughput way and a few of full-allele deletion clones are identified and confirmed by Sanger sequencing and RT-PCR (Figure C).






