Discover how semiconductor technology can create a diverse, fully covered mutant library with unbiased amino acid distribution in our application note. Download Now!
Precision Mutant Libraries
Expedite your downstream screening time with unbeatable control and complete coverage of your designed variants
Mutant libraries are powerful, high-throughput tools for optimizing protein structure and function. However, traditional methods for library construction, including error-prone PCR or degenerate methods, suffer from limited control over codon usage and poor variant representation. This requires multiple rounds of screening to capture the entire variation of the library.
Powered by GenScript's strong expertise in de novo gene synthesis and our semiconductor-based oligo synthesis technology, our Precision Mutant Library service offers precise control over each synthesized variant. The result is a more diverse and fully covered mutant library with unbiased distribution, ultimately saving valuable screening time, speeding up discovery workflow and reducing overall downstream expenses.
Customer Testimonial
"I wanted to create mutant capsids for adeno-associated virus (AAV) vectors to improve targeting and transduction levels. GenScript was able to create a combinatorial mutant library that had 100% coverage of all expected variants before and after viral packaging, and excluded variants that contained amino acids that I did not want present at these sites. Having a library with only the variants that I wanted to test reduced my screening efforts and maximized my time in finding the optimal vector sequence."
―Susan Butler, Rice University
Precision Mutant Library Options
Key Benefits of Precision Mutant Libraries
-

Precise control of user-defined codon usage

Guaranteed >90% coverage of all desired variants

Superior library diversity with library size >1010
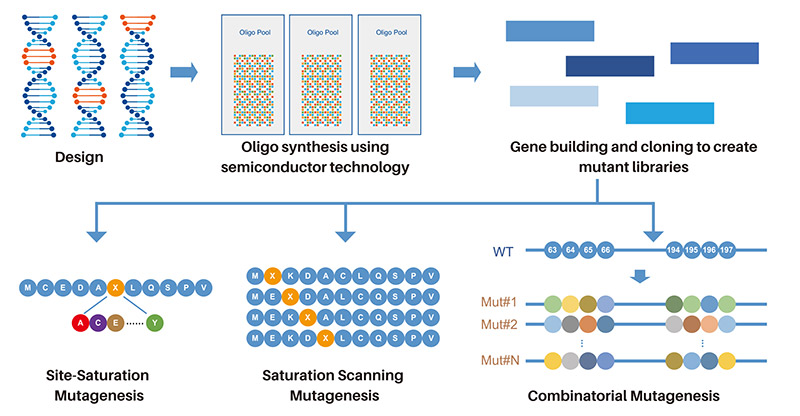
Using Next-Generation Semiconductor Technology for Library Construction
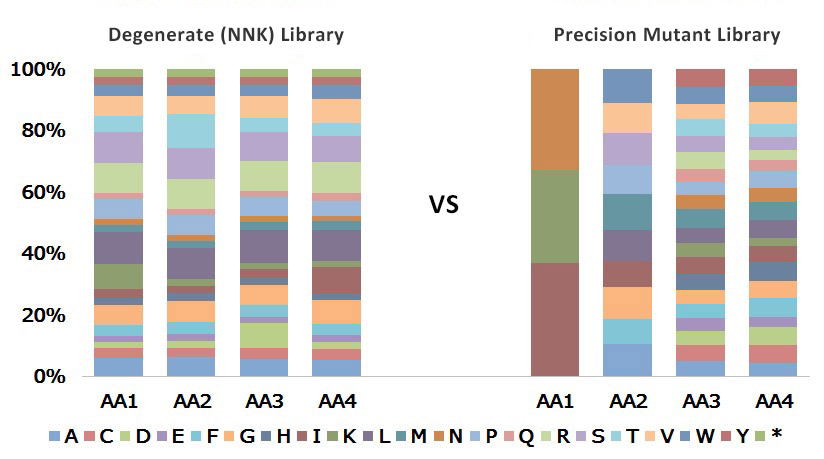
Each bar represents a mutated position and each color represents the frequency of variants with a particular amino acid.
A combinatorial mutagenesis library for 4 sequential amino acid positions was created using either degenerate (NNK) codons or using our precise oligos. Specific amino acids were requested at each site.
Using our semiconductor-based oligos in library construction:
- Eliminates the significant codon bias incurred with NNK degenerate oligos
- Achieves user-defined amino acid distribution with equal representation of variants
Service Features
| Site-Saturation Mutagenesis | Saturation Scanning Mutagenesis | Combinatorial Mutagenesis |
|---|---|---|
|
Mutate several positions one at a time within a sequence located either consecutively or scattered throughout the protein sequence.
|
Target a consecutive string of amino acids within a defined region of the sequence or the whole protein sequence.
|
Mutate multiple positions simultaneously across specific target regions of a protein.
|
Featured Resources


Feature Article in Nature:
How to make random mutagenesis less random
How to make random mutagenesis less random
Library assembly is powered by our expertise in de novo gene synthesis and advanced semiconductor technology
Mutant libraries are delivered either as a mixed pool of variants (saturation scanning or combinatorial mutagenesis libraries) or as individual subpools (site saturation mutagenesis libraries), with each pool containing all possible variants for a single amino acid position.
Cloned Libraries
- 4 µg of plasmid (high copy) for site saturation mutagenesis
- 10 µg of plasmid (high copy) for saturation scanning mutagenesis or combinatorial mutagenesis
- 1 ml of glycerol stock will be included for saturation scanning mutagenesis libraries and combinatorial mutagenesis libraries only
Double-stranded DNA (PCR Products) Libraries
- 500 ng for site saturation mutagenesis
- 4 µg for saturation scanning mutagenesis
- 500 ng-1 µg for combinatorial mutagenesis
We offer both next-generation sequencing (NGS) and Sanger sequencing to test library coverage and diversity.
- A rate over 90% of the calculated coverage for the library will be considered an acceptable library
- NGS analysis report will be provided
- Application Note: Reducing Screen Burden in Antibody Engineering Through GenScript's Synthetic Precision Libraries New Application Note: Reducing Screen Burden in Antibody Engineering Through GenScript's Synthetic Precision Libraries New
- Feature Article in Nature: How to make random mutagenesis less random Feature Article in Nature: How to make random mutagenesis less random
- Engineered antibodies: semi-rational design approach for faster optimization Engineered antibodies: semi-rational design approach for faster optimization
- (Application Note) Assembly of precise mutant libraries using arrayed-synthesized oligos for protein optimization
- (Webinar) Rapid assembly of gene variant libraries to engineer proteins for user defined goals
- (Webinar) Precision Mutant Library Helps In Protein Engineering New
A well-constructed mutant library can be a very useful tool for a variety of protein engineering projects, including:
- Improving antibody binding affinity through CDR engineering
- Optimizing enzyme affinity and stability
- Identifying critical residues within a protein domain
- Improving ligand-receptor binding
- Antibody affinity maturation
- Optimizing protein structure
How to Order
- Download and submit your mutant library design. Email the completed form to gene@genscript.com or securely upload it at Precision Mutant Library.
- Our technical account manager will contact you with a quote within 24 hours.
*For PCR product deliverables only. For more information about our rapid TAT, please contact us at gene@genscript.com
Technical Support
For questions or to discuss your library design with our Ph.D.-level scientists, contact us 24/7 via: