Novel Applications of CRISPR/Cas9 Genome Editing Technology
What is CRISPR?
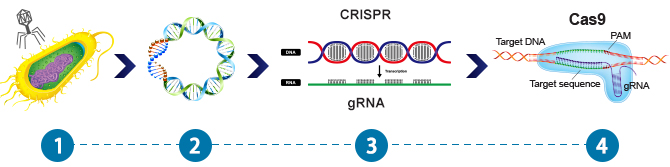
When a virus infects a bacterium, it injects its viral DNA through the bacterial cell wall (1) and into the nucleus where it becomes integrated into the host's genome (2). From there, the viral DNA will replicate become translated/transcribed into proteins which will eventually form a newly synthesized virus. In order to defend against viral infection, bacterium evolved the CRISPR/Cas9 system adaptive immune system. Within this system, bacterial CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) spacer sequences are transcribed into RNA (3) which guides a Cas9 endonuclease to matching regions of viral DNA within the hosts genome. The Cas9 endonuclease then unwinds the host cell DNA and cleaves/removes the infectious viral DNA from the bacterium (4).
Common CRISPR Applications
 CRISPR/Cas9 has become a popular gene editing tool for prokaryotic and eukaryotic systems. Due to its popularity, there are a variety of applications that CRISPR/Cas9 has been used for. One of the most common applications is using CRISPR/Cas9 for gene silencing. There are several components of the CRISPR/Cas9 system, target site specific CRISPR RNA (crRNA), trans-activating crRNA (tracrRNA) or single guide RNA (sgRNA), and Cas9 endonuclease. When a specific sequence is targeted and cleaved by the Cas9 protein within the genome, it creates a double stranded break (1). This double stranded break is repaired by the endogenous DNA repair pathway, non-homologous end joining (NHEJ). NHEJ is a mechanism that can cause errors in the genome and some of the most common errors are deletions or insertions (indels). These indels can result in frameshift mutations which can lead to premature termination of the gene or permanent silencing of the gene (2). Another common application CRISPR is used for is for transcriptional activation or repression. There have been several research groups who have developed a CRISPR/Cas9 ribonucleoprotein complex that can activate (CRISPRa) or interfere (CRISPRi) with transcription (3). CRISPR/Cas 9 can also be used in epigenetic editing where researchers can introduce epigenetic modifications into the genome (4). There are also a number of screening and imaging applications that utilize CRISPR. Below, we will discuss more applications that are recent breakthroughs and therefore a little less well known.
CRISPR/Cas9 has become a popular gene editing tool for prokaryotic and eukaryotic systems. Due to its popularity, there are a variety of applications that CRISPR/Cas9 has been used for. One of the most common applications is using CRISPR/Cas9 for gene silencing. There are several components of the CRISPR/Cas9 system, target site specific CRISPR RNA (crRNA), trans-activating crRNA (tracrRNA) or single guide RNA (sgRNA), and Cas9 endonuclease. When a specific sequence is targeted and cleaved by the Cas9 protein within the genome, it creates a double stranded break (1). This double stranded break is repaired by the endogenous DNA repair pathway, non-homologous end joining (NHEJ). NHEJ is a mechanism that can cause errors in the genome and some of the most common errors are deletions or insertions (indels). These indels can result in frameshift mutations which can lead to premature termination of the gene or permanent silencing of the gene (2). Another common application CRISPR is used for is for transcriptional activation or repression. There have been several research groups who have developed a CRISPR/Cas9 ribonucleoprotein complex that can activate (CRISPRa) or interfere (CRISPRi) with transcription (3). CRISPR/Cas 9 can also be used in epigenetic editing where researchers can introduce epigenetic modifications into the genome (4). There are also a number of screening and imaging applications that utilize CRISPR. Below, we will discuss more applications that are recent breakthroughs and therefore a little less well known.
- Jinek, M., et al., RNA-programmed genome editing in human cells. Elife, 2013. 2: p. e00471.
- Hsu, Patrick D., Eric S. Lander, and Feng Zhang. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 157.6 (2014): 1262–1278. PMC. Web. 10 Oct. 2017.
- Bikard, David, et al. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic acids research 41.15 (2013): 7429-7437.
- Liao, Jing, et al. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nature genetics 47.5 (2015): 469-478.
Novel CRISPR Applications
Endogenous Luciferase Reporter
 Common protocol to measure the affect of extracellular signals on protein expression is to transfect a cell line with a plasmid expressing a reporter (such as luciferase) under the control of known transcriptional control elements. However, this assay comes with two main constraints. First, exogenous reporter constructs are restricted to activation by only one specific signaling pathway, not allowing for other physiologically relative signaling cascades. Secondly, these reporter constructs randomly integrate into the host cell genome, leading top unknown chromatin structure. In order to circumvent these issues, researchers at Merck, have identified a way of using CRISPR/CAS9 technology to express a luciferase reporter under the control of an endogenous promoter. In order to accomplish this task, they first engineered two CRISPR plasmids, one which contained a luciferase reporter gene flanked by sequences homologous to the sequences neighboring the start codon of their gene of interest and another which containing the Cas9 and optimized guide RNA. These plasmids were co-transfected into a human leukemia cell line in order to produce a double stranded break to knockout 1 allele of the target gene while simultaneously knocking-in the luciferase gene. The final cell line contained one natural allele and one luciferase allele, both controlled under the same endogenous enhancer/promoter region. The results of the study revealed that the novel CRISPR/Cas9 endogenous luciferase reporter was able to increase luciferase activity and target protein expression in a dose dependent manor. Therefore, the authors are confident that this novel CRISPR technology will lead to much more physiological relevant expression experiments in the near future.
Common protocol to measure the affect of extracellular signals on protein expression is to transfect a cell line with a plasmid expressing a reporter (such as luciferase) under the control of known transcriptional control elements. However, this assay comes with two main constraints. First, exogenous reporter constructs are restricted to activation by only one specific signaling pathway, not allowing for other physiologically relative signaling cascades. Secondly, these reporter constructs randomly integrate into the host cell genome, leading top unknown chromatin structure. In order to circumvent these issues, researchers at Merck, have identified a way of using CRISPR/CAS9 technology to express a luciferase reporter under the control of an endogenous promoter. In order to accomplish this task, they first engineered two CRISPR plasmids, one which contained a luciferase reporter gene flanked by sequences homologous to the sequences neighboring the start codon of their gene of interest and another which containing the Cas9 and optimized guide RNA. These plasmids were co-transfected into a human leukemia cell line in order to produce a double stranded break to knockout 1 allele of the target gene while simultaneously knocking-in the luciferase gene. The final cell line contained one natural allele and one luciferase allele, both controlled under the same endogenous enhancer/promoter region. The results of the study revealed that the novel CRISPR/Cas9 endogenous luciferase reporter was able to increase luciferase activity and target protein expression in a dose dependent manor. Therefore, the authors are confident that this novel CRISPR technology will lead to much more physiological relevant expression experiments in the near future.
Liu, Jinqi, et al. "Development of a luciferase reporter Jurkat cell line under the control of endogenous interleukin-2 promoter." Journal of Immunological Methods (2017).
CasFISH: Using CRISPR for In Situ Labeling
 DNA Fluorescent In Situ Hybridization (FISH) is a widely accepted way to label specific DNA sequences. However, the standard protocol requires harmful chemicals which induce the denaturation of double stranded DNA and allow for probe hybridization, despite their known detrimental effects on biological structure and genome organization. In order to develop a simpler, less expensive, and less time consuming FISH protocol, researchers at the Transcription Imaging Consortium invented a novel way of using the CRISPR/Cas9 system to fluorescently label specific sequences of DNA. The authors first constructed a nuclease deficient Cas9 fusion protein which contained an N terminal hexahistodine tag and a C terminal halo tag bound to a JFF646 fluorophore. Next, they produced a 5'-DY547 tagged sgRNA targeting the highly repetitive mouse nucleic sequence, MajSat. Once generated, the DY547sgMajSat and the JF646-dCas9 were incubated together within mouse embryonic fibroblast in order to form a gDNA-sgRNA-dCas9 trimer. This trimer was visualized using a fluorescent microscope to show that both the DY547sgMajSat and the JF646-dCas9 co-labeled with the nuclear stain DAPI, indicating that CasFISH successfully fluorescently labeled specific DNA sequences as intended. In order to reduce the cost of this novel technology, the authors were able to successfully label their gene of interest using only fluorophore labeled Cas9, rather than also labeling the gRNA. To optimize the technique even further, the researchers identified a protocol for co-labeling several different DNA sequences at the same time by introducing multiple dCas9 proteins bound to different fluorophores simultaneously with their corresponding sgRNA's. This study was the first of its kind to not only introduce the concept of using CRISPR/Cas9 to replace DNA FISH, but also reveal its potential to replace it all together, due to CasFISH's significant reduction in cost, time, and difficulty.
DNA Fluorescent In Situ Hybridization (FISH) is a widely accepted way to label specific DNA sequences. However, the standard protocol requires harmful chemicals which induce the denaturation of double stranded DNA and allow for probe hybridization, despite their known detrimental effects on biological structure and genome organization. In order to develop a simpler, less expensive, and less time consuming FISH protocol, researchers at the Transcription Imaging Consortium invented a novel way of using the CRISPR/Cas9 system to fluorescently label specific sequences of DNA. The authors first constructed a nuclease deficient Cas9 fusion protein which contained an N terminal hexahistodine tag and a C terminal halo tag bound to a JFF646 fluorophore. Next, they produced a 5'-DY547 tagged sgRNA targeting the highly repetitive mouse nucleic sequence, MajSat. Once generated, the DY547sgMajSat and the JF646-dCas9 were incubated together within mouse embryonic fibroblast in order to form a gDNA-sgRNA-dCas9 trimer. This trimer was visualized using a fluorescent microscope to show that both the DY547sgMajSat and the JF646-dCas9 co-labeled with the nuclear stain DAPI, indicating that CasFISH successfully fluorescently labeled specific DNA sequences as intended. In order to reduce the cost of this novel technology, the authors were able to successfully label their gene of interest using only fluorophore labeled Cas9, rather than also labeling the gRNA. To optimize the technique even further, the researchers identified a protocol for co-labeling several different DNA sequences at the same time by introducing multiple dCas9 proteins bound to different fluorophores simultaneously with their corresponding sgRNA's. This study was the first of its kind to not only introduce the concept of using CRISPR/Cas9 to replace DNA FISH, but also reveal its potential to replace it all together, due to CasFISH's significant reduction in cost, time, and difficulty.
Deng, Wulan, et al. "CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells." Proceedings of the National Academy of Sciences 112.38 (2015): 11870-11875.
Gene Therapy CRISPR/Cas9 Application
 Due to its versatility, CRISPR/Cas9 genome editing tools have been used throughout different medical research areas and has shown some promising results as a potential therapy for several disease models. One method of CRISPR/Cas9 that has shown great potential in the gene therapy research area is homology directed repair (HDR). CRISPR/Cas9 is used to create a double stranded break in the DNA and ideally, one could insert a donor template to alter the coding region to potentially correct a mutation. There have been several studies discussing HDR's potential to "fix" different genetic mutations:
Due to its versatility, CRISPR/Cas9 genome editing tools have been used throughout different medical research areas and has shown some promising results as a potential therapy for several disease models. One method of CRISPR/Cas9 that has shown great potential in the gene therapy research area is homology directed repair (HDR). CRISPR/Cas9 is used to create a double stranded break in the DNA and ideally, one could insert a donor template to alter the coding region to potentially correct a mutation. There have been several studies discussing HDR's potential to "fix" different genetic mutations:
- Cong, L., et al., Multiplex genome engineering using CRISPR/Cas systems. Science, 2013. 339(6121): p. 819-23.
- Wu, Wen-Hsuan, et al. "CRISPR repair reveals causative mutation in a preclinical model of retinitis pigmentosa." Molecular Therapy 24.8 (2016): 1388-1394.
- Yin, Hao, et al. "Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo." Nature biotechnology 34.3 (2016): 328.
- Dai, Wei-Jing, et al. "CRISPR-Cas9 for in vivo gene therapy: promise and hurdles." Molecular Therapy-Nucleic Acids 5 (2016): e349.
- Kemaladewi, Dwi U., et al. "Correction of a splicing defect in a mouse model of congenital muscular dystrophy type 1A using a homology-directed-repair-independent mechanism." Nature medicine 23.8 (2017): 984.
- Yang, Su, et al. "CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington's disease." The Journal of clinical investigation 127.7 (2017).
GenScript CRISPR Genome Editing Services and Resources