| Gene Symbol | LOC373525 |
| Entrez Gene ID | 373525 |
| Full Name | MAP kinase |
| Gene Type | protein-coding |
| Organism | Strongylocentrotus purpuratus(purple sea urchin) |
-
REAGENT SERVICES
Hot!
-
Most Popular Services
-
Molecular Biology
-
Recombinant Antibody/Protein
-
Reagent Antibody
-
CRISPR Gene Editing
-
DNA Mutant Library
-
IVT RNA and LNP Formulations
-
Oligo Synthesis
-
Peptides
-
Cell Engineering
-
- Gene Synthesis FLASH Gene
- GenBrick™ Up to 200kb
- Gene Fragments Up to 3kb now
- Plasmid DNA Preparation Upgraded
- Cloning and Subcloning
- ORF cDNA Clones
- mRNA Plasmid Solutions New!
- Cell free mRNA Template New!
- AAV Plasmid Solutions New!
- Mutagenesis
- GenCircle™ Double-Stranded DNA New!
- GenSmart™ Online Tools
-
-
PRODUCTS
-
Most Popular Reagents
-
 Instruments
Instruments
-
Antibodies
-
ELISA Kits
-
Protein Electrophoresis and Blotting
-
Protein and Antibody Purification
-
Recombinant Proteins
-
Molecular Biology
-
Stable Cell Lines
-
Cell Isolation and Activation
-
 IVD Raw Materials
IVD Raw Materials
-
 Therapy Applications
Therapy Applications
-
Resources
-
- All Instruments
- Automated Protein and Antibody Purification SystemNew!
- Automated Plasmid MaxiprepHot!
- Automated Plasmid/Protein/Antibody Mini-scale Purification
- eBlot™ Protein Transfer System
- eStain™ Protein Staining System
- eZwest™ Lite Automated Western Blotting Device
- CytoSinct™ 1000 Cell Isolation Instrument
-
- Pharmacokinetics and Immunogenicity ELISA Kits
- Viral Titration QC ELISA Kits
- -- Lentivirus Titer p24 ELISA KitHot!
- -- MuLV Titer p30 ELISA KitNew!
- -- AAV2 and AAVX Titer Capsid ELISA Kits
- Residual Detection ELISA Kits
- -- T7 RNA Polymerase ELISA KitNew!
- -- BSA ELISA Kit, 2G
- -- Cas9 ELISA KitHot!
- -- Protein A ELISA KitHot!
- -- His tagged protein detection & purification
- dsRNA ELISA Kit
- Endonuclease ELISA Kit
- COVID-19 Detection cPass™ Technology Kits
-
- Automated Maxi-Plasmid PurificationHot!
- Automated Mini-Plasmid PurificationNew!
- PCR Reagents
- S.marcescens Nuclease Benz-Neburase™
- DNA Assembly GenBuilder™
- Cas9 / Cas12a / Cas13a Nucleases
- Base and Prime Editing Nucleases
- GMP Cas9 Nucleases
- CRISPR sgRNA Synthesis
- HDR Knock-in Template
- CRISPR Gene Editing Kits and Antibodies
-
![AmMag™ Quatro Automated Plasmid Purification]() AmMag™ Quatro automated plasmid purification
AmMag™ Quatro automated plasmid purification
-
![Anti-Camelid VHH]() MonoRab™ Anti-VHH Antibodies
MonoRab™ Anti-VHH Antibodies
-
![ELISA Kits]() ELISA Kits
ELISA Kits
-
![Precast Gels]() SurePAGE™ Precast Gels
SurePAGE™ Precast Gels
-
![Quatro ProAb Automated Protein and Antibody Purification System]() AmMag™ Quatro ProAb Automated Protein and Antibody Purification System
AmMag™ Quatro ProAb Automated Protein and Antibody Purification System
-
![Target Proteins]() Target Proteins
Target Proteins
-
![AmMag™ Quatro Automated Plasmid Purification]() AmMag™ Quatro automated plasmid purification
AmMag™ Quatro automated plasmid purification
-
![Stable Cell Lines]() Stable Cell Lines
Stable Cell Lines
-
![Cell Isolation and Activation]() Cell Isolation and Activation
Cell Isolation and Activation
-
 IVD Raw Materials
IVD Raw Materials
-
![Quick
Order]() Quick Order
Quick Order
-
![Quick
Order]() Quick Order
Quick Order
- APPLICATIONS
- RESOURCES
- ABOUT US
- SIGN IN My Account SIGN OUT
- REGISTER
ORF » Species Summary » Strongylocentrotus purpuratus » LOC373525 cDNA ORF clone
LOC373525 cDNA ORF clone, Strongylocentrotus purpuratus(purple sea urchin)
-
Gene
-
Clones
gRNAs
-
Summary
-
mRNA and Protein(s)
mRNA Protein Name XM_011684177.1 XP_011682479.1 MAP kinase isoform X1 NM_214648.1 NP_999813.1 MAP kinase -
Pathways from BioSystems

-
Gene Ontology

-
Bibliography
Related articles in PubMed
Gene structure in the sea urchin Strongylocentrotus purpuratus based on transcriptome analysis.
Tu Q, Cameron RA, Worley KC, Gibbs RA, Davidson EH
Genome research22(10)2079-87(2012 Oct)GeneRIFs: Gene References Into Functions What's a GeneRIF?
The following LOC373525 gene cDNA ORF clone sequences were retrieved from the NCBI Reference Sequence Database (RefSeq). These sequences represent the protein coding region of the LOC373525 cDNA ORF which is encoded by the open reading frame (ORF) sequence. ORF sequences can be delivered in our standard vector, pcDNA3.1+/C-(K)DYK or the vector of your choice as an expression/transfection-ready ORF clone. Not the clone you want? Click here to find your clone.
-
Reference Sequences (Refseq)
CloneID OSd20236 Clone ID Related Accession (Same CDS sequence) NM_214648.1 Accession Version NM_214648.1 Latest version! Documents for ORF clone product in default vector Sequence Information ORF Nucleotide Sequence (Length: 1110bp)
Protein sequence
SNPVector pcDNA3.1-C-(k)DYK or customized vector  User Manual
User ManualClone information Clone Map  MSDS
MSDSTag on pcDNA3.1+/C-(K)DYK C terminal DYKDDDDK tags ORF Insert Method CloneEZ™ Seamless cloning technology Insert Structure linear Update Date 1569600000000 Organism Strongylocentrotus purpuratus(purple sea urchin) Product MAP kinase Comment Comment: PROVISIONAL REFSEQ: This record has not yet been subject to final NCBI review. The reference sequence was derived from AY145443.1. On Aug 12, 2005 this sequence version replaced XM_781270.1. ##Evidence-Data-START## Transcript exon combination :: AY145443.1 [ECO:0000332] RNAseq introns :: mixed/partial sample support SAMN01103187, SAMN01103196 [ECO:0000350] ##Evidence-Data-END##
1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081ATGGCCGACC AAGGAGGTGC AAAAACTAAG CGGACCAACG AAGAGAAACC TGAAACTGTG
AGAGGACAAG TCTTCGATGT CGGTCCACGC TATGTTACCC TGAACTACAT CGGGGAAGGG
GCTTATGGAA TGGTTTGTAG TGCGGTGGAT ACACGACACG GAGGCAAGGT GGCTATCAAG
AAGATCAGTC CGTTTGAGCA CCAGACATAC TGCCAACGAA CCCTCAGAGA GATTAAAATC
CTTACACGCT TCAATCATGA AAATATTATT AATATCCAAG ATATCATCAA GGCAGACACA
ATTGAAGCCA TGAGAGATGT ATATATTGTT CAAAGTCTAA TGGAAACAGA CCTGTACAAA
CTGCTCAAGA CCCAGCCGCT GAGCAACGAC CACATCTGCT ACTTCCTCTA CCAGATCCTT
CGAGGGCTCA AGTACATCCA CTCGGCCAAC GTGCTTCACA GGGACCTCAA ACCAAGCAAC
CTGCTGCTCA ATACTACCTG TGATCTCAAG ATCTGCGACT TTGGACTTGC ACGGATAGCT
GACCCCGGCC ACGACCACAC AGGTTTCTTG ACGGAATACG TAGCGACGCG ATGGTACCGC
GCCCCCGAGA TCATGCTGAA TTCCAAGGGT TACACCAAGT CCATTGACAT CTGGTCGGTC
GGTTGCATTC TGGCTGAGAT GTTGAGCAAC AGACCCATCT TTCCTGGAAA ACATTATCTG
GATCAGTTGA ACCACATCCT GGGAGTTCTA GGTTCGCCAT CCCAAGACGA CCTGAAATGC
ATCATCAACG AGAAGGCCAG AGCCTACCTC CAGGGACTAC CGTTCAAGTC CAAGATTCCC
CTCAAATCAC TCTTCCCGAA GGCTGATAAC AAAGCTCTTG ATTTCTTGGA GAGGATGCTT
TCCTTCAACC CTGACAAGAG GATAACTGTA GAGGAAGCTC TAGCACATCC CTACCTGGAA
CAATACTATG ATCCAGATGA TGAGCCTGTC AAAGAAGAGC CGTTCACATT TGTCACGGAG
CTGGATGATT TACCAAAGGA GAAACTGAAA GAGATGATCT TTGAAGAAGC CAGTAAATTC
AACCCATCCC CAAGCCCAAA TACTAGCTGAThe stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
RefSeq NP_999813.1 CDS 436..1545 Translation 
Target ORF information:
RefSeq Version NM_214648.1 Organism Strongylocentrotus purpuratus(purple sea urchin) Definition Strongylocentrotus purpuratus MAP kinase (LOC373525), mRNA. Target ORF information:
Epitope DYKDDDDK Bacterial selection AMPR Mammalian selection NeoR Vector pcDNA3.1+/C-(K)DYK 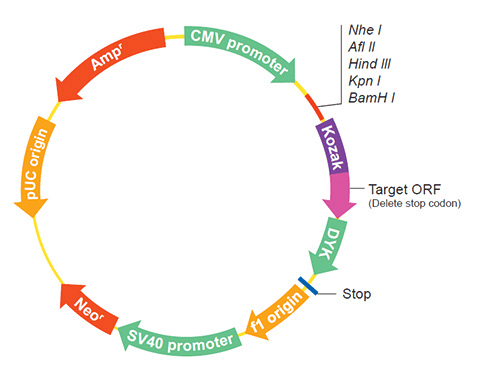 NM_214648.1
NM_214648.1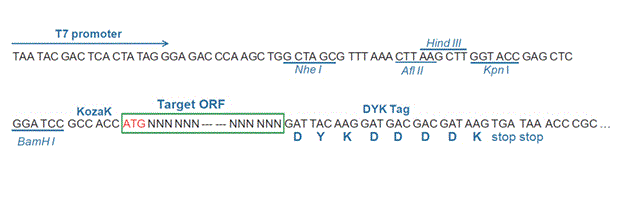
ORF Insert Sequence:
1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081ATGGCCGACC AAGGAGGTGC AAAAACTAAG CGGACCAACG AAGAGAAACC TGAAACTGTG
AGAGGACAAG TCTTCGATGT CGGTCCACGC TATGTTACCC TGAACTACAT CGGGGAAGGG
GCTTATGGAA TGGTTTGTAG TGCGGTGGAT ACACGACACG GAGGCAAGGT GGCTATCAAG
AAGATCAGTC CGTTTGAGCA CCAGACATAC TGCCAACGAA CCCTCAGAGA GATTAAAATC
CTTACACGCT TCAATCATGA AAATATTATT AATATCCAAG ATATCATCAA GGCAGACACA
ATTGAAGCCA TGAGAGATGT ATATATTGTT CAAAGTCTAA TGGAAACAGA CCTGTACAAA
CTGCTCAAGA CCCAGCCGCT GAGCAACGAC CACATCTGCT ACTTCCTCTA CCAGATCCTT
CGAGGGCTCA AGTACATCCA CTCGGCCAAC GTGCTTCACA GGGACCTCAA ACCAAGCAAC
CTGCTGCTCA ATACTACCTG TGATCTCAAG ATCTGCGACT TTGGACTTGC ACGGATAGCT
GACCCCGGCC ACGACCACAC AGGTTTCTTG ACGGAATACG TAGCGACGCG ATGGTACCGC
GCCCCCGAGA TCATGCTGAA TTCCAAGGGT TACACCAAGT CCATTGACAT CTGGTCGGTC
GGTTGCATTC TGGCTGAGAT GTTGAGCAAC AGACCCATCT TTCCTGGAAA ACATTATCTG
GATCAGTTGA ACCACATCCT GGGAGTTCTA GGTTCGCCAT CCCAAGACGA CCTGAAATGC
ATCATCAACG AGAAGGCCAG AGCCTACCTC CAGGGACTAC CGTTCAAGTC CAAGATTCCC
CTCAAATCAC TCTTCCCGAA GGCTGATAAC AAAGCTCTTG ATTTCTTGGA GAGGATGCTT
TCCTTCAACC CTGACAAGAG GATAACTGTA GAGGAAGCTC TAGCACATCC CTACCTGGAA
CAATACTATG ATCCAGATGA TGAGCCTGTC AAAGAAGAGC CGTTCACATT TGTCACGGAG
CTGGATGATT TACCAAAGGA GAAACTGAAA GAGATGATCT TTGAAGAAGC CAGTAAATTC
AACCCATCCC CAAGCCCAAA TACTAGCTGAThe stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
CloneID OSd20607 Clone ID Related Accession (Same CDS sequence) XM_011684177.1 Accession Version XM_011684177.1 Latest version! Documents for ORF clone product in default vector Sequence Information ORF Nucleotide Sequence (Length: 1110bp)
Protein sequence
SNPVector pcDNA3.1-C-(k)DYK or customized vector  User Manual
User ManualClone information Clone Map  MSDS
MSDSTag on pcDNA3.1+/C-(K)DYK C terminal DYKDDDDK tags ORF Insert Method CloneEZ™ Seamless cloning technology Insert Structure linear Update Date 1427040000000 Organism Strongylocentrotus purpuratus(purple sea urchin) Product MAP kinase isoform X1 Comment Comment: MODEL REFSEQ: This record is predicted by automated computational analysis. This record is derived from a genomic sequence (NW_011995189.1) and transcript sequence (AY145443.1) annotated using gene prediction method: Gnomon, supported by mRNA and EST evidence. Also see: Documentation of NCBI's Annotation Process ##Genome-Annotation-Data-START## Annotation Provider :: NCBI Annotation Status :: Full annotation Annotation Version :: Strongylocentrotus purpuratus Annotation Release 101 Annotation Pipeline :: NCBI eukaryotic genome annotation pipeline Annotation Software Version :: 6.2 Annotation Method :: Best-placed RefSeq; Gnomon Features Annotated :: Gene; mRNA; CDS; ncRNA ##Genome-Annotation-Data-END## ##RefSeq-Attributes-START## assembly gap :: added 40 transcript bases to patch genome assembly gap ##RefSeq-Attributes-END##
1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081ATGGCCGACC AAGGAGGTGC AAAAACTAAG CGGACCAACG AAGAGAAACC TGAAACTGTG
AGAGGACAAG TCTTCGATGT CGGTCCACGC TATGTTACCC TGAACTACAT CGGGGAAGGG
GCTTATGGAA TGGTTTGTAG TGCGGTGGAT ACACGACACG GAGGCAAGGT GGCTATCAAG
AAGATCAGTC CGTTTGAGCA CCAGACATAC TGCCAACGAA CCCTCAGAGA GATTAAAATC
CTTACACGCT TCAATCATGA AAATATTATT AATATCCAAG ATATCATCAA GGCAGACACA
ATTGAAGCCA TGAGAGATGT ATATATTGTT CAAAGTCTAA TGGAAACAGA CCTGTACAAA
CTGCTCAAGA CCCAGCCGCT GAGCAACGAC CACATCTGCT ACTTCCTCTA CCAGATCCTT
CGAGGGCTCA AGTACATCCA CTCGGCCAAC GTGCTTCACA GGGACCTCAA ACCAAGCAAC
CTGCTGCTCA ATACTACCTG TGATCTCAAG ATCTGCGACT TTGGACTTGC ACGGATAGCT
GACCCCGGCC ACGACCACAC AGGTTTCTTG ACGGAATACG TAGCGACGCG ATGGTACCGC
GCCCCTGAGA TCATGCTGAA TTCCAAGGGT TACACCAAGT CCATTGACAT CTGGTCGGTT
GGTTGCATTC TGGCCGAGAT GTTGAGCAAC AGACCCATCT TTCCTGGAAA ACATTATCTG
GATCAGCTGA ACCACATCCT GGGAGTTCTA GGTTCGCCAT CTCAGGACGA CCTGAAGTGC
ATCATCAACG AGAAGGCCAG AGCCTACCTC CAGGGACTAC CCTTCAAGTC CAAGATTCCC
CTCAAATCAC TCTTCCCGAA GGCTGATAAC AAAGCTCTTG ATTTCTTGGA GAGGATGCTT
TCCTTCAACC CTGACAAGAG GATAACTGTA GAGGAAGCTC TAGCACATCC CTACCTGGAA
CAATACTATG ATCCAGATGA TGAGCCTGTC AAAGAAGAGC CGTTCACATT TGTCACGGAG
CTGGATGATT TACCAAAGGA GAAACTGAAA GAGATGATCT TTGAAGAAGC CAGTAAATTC
AACCCATCCC CAAGCCCCAA TACTAGCTGAThe stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
RefSeq XP_011682479.1 CDS 393..1502 Translation 
Target ORF information:
RefSeq Version XM_011684177.1 Organism Strongylocentrotus purpuratus(purple sea urchin) Definition Strongylocentrotus purpuratus MAP kinase (LOC373525), transcript variant X1, mRNA. Target ORF information:
Epitope DYKDDDDK Bacterial selection AMPR Mammalian selection NeoR Vector pcDNA3.1+/C-(K)DYK  XM_011684177.1
XM_011684177.1
ORF Insert Sequence:
1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081ATGGCCGACC AAGGAGGTGC AAAAACTAAG CGGACCAACG AAGAGAAACC TGAAACTGTG
AGAGGACAAG TCTTCGATGT CGGTCCACGC TATGTTACCC TGAACTACAT CGGGGAAGGG
GCTTATGGAA TGGTTTGTAG TGCGGTGGAT ACACGACACG GAGGCAAGGT GGCTATCAAG
AAGATCAGTC CGTTTGAGCA CCAGACATAC TGCCAACGAA CCCTCAGAGA GATTAAAATC
CTTACACGCT TCAATCATGA AAATATTATT AATATCCAAG ATATCATCAA GGCAGACACA
ATTGAAGCCA TGAGAGATGT ATATATTGTT CAAAGTCTAA TGGAAACAGA CCTGTACAAA
CTGCTCAAGA CCCAGCCGCT GAGCAACGAC CACATCTGCT ACTTCCTCTA CCAGATCCTT
CGAGGGCTCA AGTACATCCA CTCGGCCAAC GTGCTTCACA GGGACCTCAA ACCAAGCAAC
CTGCTGCTCA ATACTACCTG TGATCTCAAG ATCTGCGACT TTGGACTTGC ACGGATAGCT
GACCCCGGCC ACGACCACAC AGGTTTCTTG ACGGAATACG TAGCGACGCG ATGGTACCGC
GCCCCTGAGA TCATGCTGAA TTCCAAGGGT TACACCAAGT CCATTGACAT CTGGTCGGTT
GGTTGCATTC TGGCCGAGAT GTTGAGCAAC AGACCCATCT TTCCTGGAAA ACATTATCTG
GATCAGCTGA ACCACATCCT GGGAGTTCTA GGTTCGCCAT CTCAGGACGA CCTGAAGTGC
ATCATCAACG AGAAGGCCAG AGCCTACCTC CAGGGACTAC CCTTCAAGTC CAAGATTCCC
CTCAAATCAC TCTTCCCGAA GGCTGATAAC AAAGCTCTTG ATTTCTTGGA GAGGATGCTT
TCCTTCAACC CTGACAAGAG GATAACTGTA GAGGAAGCTC TAGCACATCC CTACCTGGAA
CAATACTATG ATCCAGATGA TGAGCCTGTC AAAGAAGAGC CGTTCACATT TGTCACGGAG
CTGGATGATT TACCAAAGGA GAAACTGAAA GAGATGATCT TTGAAGAAGC CAGTAAATTC
AACCCATCCC CAAGCCCCAA TACTAGCTGAThe stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
-
PubMed
Gene structure in the sea urchin Strongylocentrotus purpuratus based on transcriptome analysis.
Genome research22(10)2079-87(2012 Oct)
Tu Q,Cameron RA,Worley KC,Gibbs RA,Davidson EH
-
-
Most Popular Services
-
Services & Products
-
News & Blogs
- Measles Attack: Ultra-Fast Virus Detection by Peptide Libraries
- Identifying New Anti-PD-1 Monoclonal Antibody Drug Candidates
- Gene variant found in millions of Americans could result in obesity
- Studies show that the RBD is the best antigen to induce neutralizing antibodies for COVID-19
- The combination of CDK4/6 inhibitor and PD-1/PD-L1 immune checkpoint blocker is expected to enhance treatment efficacy
- Combinatorial vs Sequential Optimization for Metabolic Pathway Engineering, What Are The Advantages?
- Formation of flat surfaces critical in helping macrophages eat
- Dig gold out of “junks”
-
-
-
PubMed
 Hi!Ask me about GenScript services and products! I can answer questions or connect you to a live person.
Hi!Ask me about GenScript services and products! I can answer questions or connect you to a live person. -




































